













| General Information: |
|
| Name(s) found: |
l(3)82Fd-PF /
FBpp0078456
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 869 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell wall macromolecule catabolic process
[IEA]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHTAVFWRNG DILEGLRPGS PKPGHIERVG GSSQQQQAEE EANKSDDPVI TQRFLKINVR 60
61 HITDGQGVVG GVLLVTPNAV MFDPNVSDPL VIEHGPESYG VIAPMELVVN AAIFHDIAHM 120
121 RVAGGAGQGL NSASGDAEKP EIYYPKPVLV EEDSKELPEQ QSLLGEDGNK RLDAEIGSLE 180
181 IADDQESLCS STGRDGDAFP KAFDRERVED SSMEAKDSAK TDALDDEDKK AGLGLSSTRS 240
241 TLEERRKSLL DHHWAIPSKD RSSEDEGDNE SNVTVDSGAR VQDPHSANSS VSGVPLSQPP 300
301 VAAAAAAAAV APASAVVPLP PSGIDLEHLE QLSKQSCYDS GIDIREPVPN VQPIPKKTVY 360
361 SDADIVLSSD WVPPKTIVPT HFSESPPRST ILGQSLDAGT GGAGGVRKKT SSVSFSVDDE 420
421 AAQQAQAQSA ALAVTDKQAD KKNKMLKRLS YPLTWVEGLT GEGGAVPPGG VGSGSLNKSG 480
481 DTDSAPNTGD SNQSVFSKVF SRRSSIGTFI RPHSSEGTSS STKLKEAKQA PKLDYRSMVS 540
541 MDDKPELFIS VDKLIPRPAR ACLDPPLYLR LRMGKPIGKA IPLPTSVMSY GKNKLRAEYW 600
601 FSVPKNRVDE LYRFINTWVK HLYGELDEEQ IKARGFELIQ EDTEWTKSGT TKAGMGGGSQ 660
661 DGEEISDLTR ESWEVLSMST DEYRKTSLFA TGSFDLDFPI PDLIGKTEIL TEEHREKLCS 720
721 HLPARAEGYS WSLIFSTSQH GFALNSLYRK MARLESPVLI VIEDTEHNVF GALTSCSLHV 780
781 SDHFYGTGES LLYKFNPSFK VFHWTGENMY FIKGNMESLS IGAGDGRFGL WLDGDLNQGR 840
841 SQQCSTYGNE PLAPQEDFVI KTLECWAFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
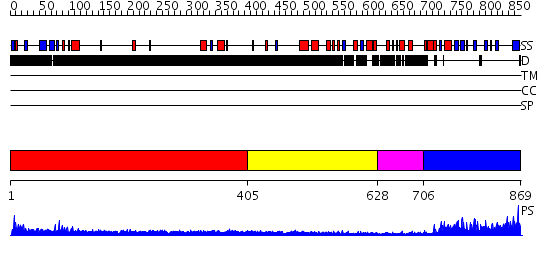
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..404] | 4.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [405..627] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [628..705] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [706..869] | 75.823909 | No description for PF07534.7 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)