













| General Information: |
|
| Name(s) found: |
gi|62484392
[NCBI NR]
gi|61678332 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 812 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 PNRLSGITFR SSPQRLLIGL ETQPTANVGS PSPLQNQRFR VRDMDNEQIM STFGNIGHLL 60
61 NIAVERFITI GEIIAEENID IKGDMYEAAK EARDAGKSIE RLCDISPLSG MELRHHIEPL 120
121 KDYGAIILAA RSLLSSVTRI LLLVDIIVVK KLLTAKKRAS ESLEKLESVM NFTEFVRAFS 180
181 VFGTEMIELA YLTGHHRNSF KEERRRAQMF SARQILEKSI AILLTSSKNS LIHSDCVIVK 240
241 ENRDTVFCQI RRAMDLIHFV VKDSIFDTAK HMPFEKQASN PNYENESIYT TIKRIVNLIK 300
301 RYKYQMSYYE GSGEGNNNSD AYFSFLDDDL WKSEKNLPSA AGGGRMVNNT NFRKHLSSES 360
361 LDLREELSLA FEKLFEKAHD FTDSPYISHN HRKNILSYCD NCKLEFDLYL NTIMKDQHEG 420
421 GGGAGGVGGV GGGQVKGRSD PELGMLSSLE ELCKQLVVSV SSQIEDFNES LKICSKLVKS 480
481 FHVLATNYDI DNLNQRSSKF HDCCDHIFDI CKLLQHIAPT ERLQVQAKCL AINLRIYGPQ 540
541 VFIAAKILWK YSNSSAANEN FESFSDMWKW LTNEISMIAK KILVSINTTK GDRDADAEKC 600
601 ESSSDKLTKS GVPATEDGEM VEYSGLEGKN NGDLNTYQSK WSEDLSDDND ILKRAKNMSA 660
661 MAFLMYQFTK GNGSLRTTQD LFTQAEYFAE EANRLYKVLR HFSYQVPASD NKKDLLNILD 720
721 RVPTFVQALQ FTVKDHTVGK AATFVKVDHV IRETKNLMNV INKVVSKCFE CATKYKLNLT 780
781 GITGGLTSER SNENMAGGMC DSKGTTSSSD IP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
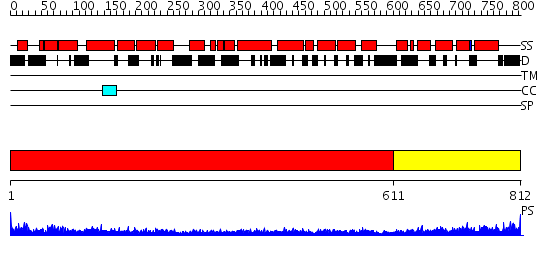
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..610] | 60.154902 | Crystal structure of human full-length vinculin (residues 1-1066) | |
| 2 | View Details | [611..812] | 24.69897 | Human vinculin head (1-258) in complex with human vinculin tail (879-1066) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)