













| General Information: |
|
| Name(s) found: |
a-PC /
FBpp0099961
[FlyBase]
a-PB / FBpp0071678 [FlyBase] a-PA / FBpp0071677 [FlyBase] |
| Description(s) found:
Found 50 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1329 amino acids |
Gene Ontology: |
|
| Cellular Component: |
adherens junction
[IDA][NAS]
apical plasma membrane [IDA] |
| Biological Process: |
compound eye development
[IMP]
|
| Molecular Function: |
protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLFKTRKST DTYSTLAAQQ QQQQQQQQQH QAEGSNISHS SNSSSNKSHT PATCSNRLNK 60
61 SIVSSTSISS SLPDLHDKSP VMILSCTTLA SNGATATAAV TATATGTAAT SGGSLQQQQQ 120
121 QHLQHQQQQQ PLRTATPTCL LSGRQTPSAI SVMSLQEATS LHRQQQQPHQ PPTIYVPVPT 180
181 KLGNNVNTGN SSATLLLSYG STSSIANLQQ QQQQHAAQYQ QYVAQRLHAA SSSCLYEKGS 240
241 NASGGASSNK SSLSLTPNGH LPDYKLVTAM PVVVLDDEHK SNSLPATEAS RNSNSSSNMN 300
301 GSSNSNSLDV SNSNSHSGSS TSLASTTRNV FTWGKRMSRK LDLLKRSDSP AAAHKSHSDL 360
361 RSLFHSPTHH KSGSGGSSGP SSAKASASPT GGHQNSSGST TSTLKKCKSG PIETIKQRHQ 420
421 QQQQQQQSVQ DVGTGQSQSA QSTPTHQFQA AARPQKALKN FFHRIGSTGM LNHRSHNLLK 480
481 ASEAAQQATP AATTLYRSSS TSQLSSSSYV KCDDPTEGLN LQREQREQRL PRIASLKSSS 540
541 CDDIAKVSSC LTASTSSGSA AGSLGSPPSS AAAGGGGTAN SGQHDPSRRG AFPYAFLRSR 600
601 LSVLPEENHG NVPGHLKQQI QRQREQHQQH QRDLLQQEQT SSPLPQRRSP EQAMLNNVSR 660
661 NDSITSKDWE PLYQRLSSCL SSNESGYDSD GGATGARLGN NLSISGGDTE SIASGTLKRN 720
721 SLISLSSSEG VGMGMGMSLG LGAPSTRNSS ICSAPVSLGG YNYDYETETI RRRFRQVKLE 780
781 RKCQEDYIGI VLSPKTVMTN SNEQQYRYLI VELEPYGMAQ KDGRLRLGDE IVNVNGKHLR 840
841 GIQSFAEVQR LLSSFVDNCI DLVIAHDEVT TVTDFYTKIR IDGMSTQRHR LSYVQRTQST 900
901 DSLSSMQSLQ LQQERIQGHN TEQEQEAQGE DQCDARSMAS VSTMPTPMPL MQHRRSSTPR 960
961 HSLDVGAPEH ELLRRRARSS SGQRSLALTP TPLFASGSSS CSSSPNHRLL DNENDPANDT1020
1021 DSYTPVYANR AASVCVASSL ADDEKWQLLA RKRCSEGSAL SATPNPQQFG QRTHYARNSI1080
1081 NLANSHYRSL RFAHSRLSSS RLSLFMQAPP NSLTVGEGVA NTPSSTATTT TDLTNQQQQQ1140
1141 QNQQQTHQSL YIKHSPKSVS LFSPNPYVNA SSSPASASTS AGAGSSLAPP AAALMHHRPS1200
1201 LPVAKLTIRD EEMAEVIRAS MSEGSGRCTP KTITFFKGPG LKSLGFSIVG GRDSPKGNMG1260
1261 IFVKTVFPSG QAADDGTLQA GDEIVEINGN SVQGMSHAET IGLFKNVREG TIVLKILRRK1320
1321 LQKAKSMGC |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
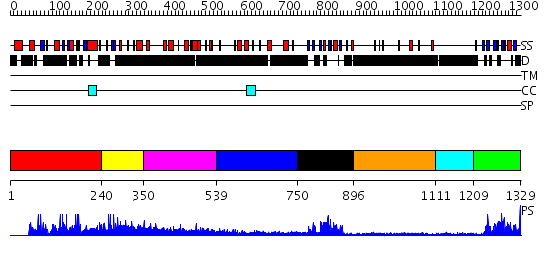
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..239] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [240..349] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [350..538] | 2.135989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [539..749] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [750..895] | 16.69897 | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein) | |
| 6 | View Details | [896..1110] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1111..1208] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1209..1329] | 27.522879 | The crystal structure of the 13th PDZ domain of MPDZ |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 |
|
|||||||||||||||
| 6 | No functions predicted. | |||||||||||||||
| 7 | No functions predicted. | |||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)