













| General Information: |
|
| Name(s) found: |
PTP-ER-PA /
FBpp0071603
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1377 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
inactivation of MAPK activity
[NAS]
protein amino acid dephosphorylation [IDA][NAS] R7 cell fate commitment [IGI] Ras protein signal transduction [IGI] mitotic cell cycle [IMP] |
| Molecular Function: |
protein tyrosine phosphatase activity
[IDA][ISS][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKLRNNNKE NAPIEYPYPQ TPRPTHAAPS DTTPLEAGED AESNKSTPYF TPLEYLQTSF 60
61 EFPSPTVEVG GGVPGFPEHE DQEEEVFTPR QFTITSPHPA AHVHHPSSSQ SPTLGRKFKR 120
121 LSQYDRRSCS PHINTSSGEE RVSGRGIDKE NTMPKGGSVD QDLENSLYLS KSEHNSPTIL 180
181 FKDESSTTPG LEASRLSYSP KLLSSINNLN VSGSPLNIIN SAGYKNGNFF RFPEIDHVVQ 240
241 DAPMDTVCLE EATRRDRSPA LAEPPEQQQQ KSEGRKNRKN KNNLTIRITD VPIKNSSQPS 300
301 PLPKPKTPTI KSTKEKARSL DSAANESELS IVVHNITESH GSCDDMETGQ SQPSTSVIHH 360
361 HLHLQQPASA IHSTSLSRLD SHLSLANQRR TPRRKSPGIN TTDWLMYHRK QNPYQVQPTH 420
421 CSSTTQSSLD SDASLTPSLG DFELKSACSV DGGSKFGIGA SLAPRSAHKH NQLLHSSSTN 480
481 LKTLPECLTL VEFSSSGGGP KESPFKQKSM DLPMPTLQAK TTVSTSSMNL LQRRGSNHSL 540
541 TLNLHSSCGN LMNSGLSHSN YSLGSILNSK SSCNLDAGTA SQIRADQLPQ SHLLGSSQGS 600
601 RQGLFRRRGS NHSITLKTGH TTTSCGDLNS MQQLQQQQAM LSAEKYNRLN MRTNSTGSGS 660
661 GSGSNVPTPK RGLLERRGSN QSLTLNMGSI LGGAAGTEIQ MTDIVQPKVT VCSCAAANQA 720
721 PHQRKFFSTE NLNSSKANSQ KFFGQVCFGS ASDLQPNPQQ DSVREDQAIG ATCTCAGTVR 780
781 NIMTRPLSPQ TTSEEFKIYL ASIQMLQSAS NPLNQFDLIR LNYVFDHSYQ TNRNIIEQPP 840
841 VLGSNARDVE TPTDMVPPSS EDSELLASYQ TVVSRMAQPI PELKENEQKR IFRDLHKEFW 900
901 DLPLNHQEKP MVFGSQTKNR YKTILPNENS RVLLESESSE LTSLLGEIKR TSSVTASEDL 960
961 PYINANYIKG PDYVSKCYVA TQGPLPNTIF EFWLMIYQNT QRYIRRCVDG GSSSSPHVDR1020
1021 EQILQQYFQK IVMLTNFTEA NRQKCAVYFP IELNEIFAVA AKCEVFQLSA AARDYFDRYL1080
1081 TPTFVPDTVV ASSDAIDYEI SGRHIGVESV KVTLEGDLLE ALPAQGSFFL IKNVGIVRRN1140
1141 GYSVRKLVLL YCIRVPQSAS YHLQKIYCYH YWYPDWPDHH SPRDINTLLD TCLHVLNLGK1200
1201 CESEFDIYDD TRSERNAHLA AQRLEIYQQD IFNAVQPLPV IHCSAGIGRT GCFTAILNAV1260
1261 RQLRQSLAYS LTGMLTKSLT SSSTEEYHNP TDSDSSFTCN TIRHISHILD HRDAEAVKTP1320
1321 PSFDRLPKMP DIFVDVLGIV CNLRLQRGGM VQNSEQYELI HRAICLYLKR TLALRRF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
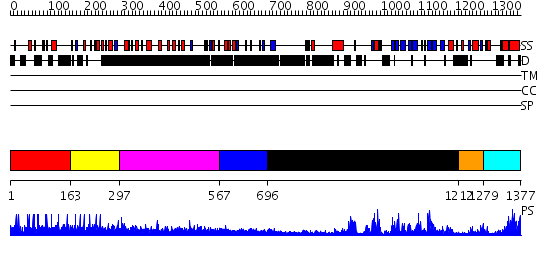
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [163..296] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [297..566] | 4.148997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [567..695] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [696..1211] | 90.30103 | Crystal structure of human tyrosine phosphatase SHP-1 | |
| 6 | View Details | [1212..1278] | 89.39794 | Crystal structure of the tandem phosphatase domain of RPTP CD45 | |
| 7 | View Details | [1279..1377] | 122.0 | Crystal structure of the phosphatase domains of human PTP SIGMA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)