













| General Information: |
|
| Name(s) found: |
insc-PA /
FBpp0071490
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apical part of cell
[NAS]
cytoplasm [IDA] cell cortex [IDA] apical cortex [IDA][TAS] |
| Biological Process: |
cytoskeleton organization
[NAS]
establishment of mitotic spindle localization [IMP][NAS][TAS] protein localization [IMP][NAS] peripheral nervous system development [IMP] neuroblast fate determination [TAS] asymmetric cell division [TAS] somatic muscle development [IMP] RNA localization [IMP] sensory organ development [TAS] asymmetric protein localization [TAS] asymmetric protein localization involved in cell fate determination [NAS][TAS] basal protein localization [TAS] apical protein localization [IMP][IPI] asymmetric neuroblast division [IGI][TAS] |
| Molecular Function: |
cytoskeletal adaptor activity
[IMP][ISS][NAS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFQRSYSKV WWGSEQSSLT SGGFYSSFSG GSSTSSTSNY PQRQPASLGP LGNLSCIQEV 60
61 EDEQNSSKLS WHKHDSPGSL RSQDSGFSDN EESHQLQHQQ NLSPPSPQSD KQQTPAATPT 120
121 SVHSVATPPT VVRRVGNSSF YSPLISVTRR ISFTSGQITP KTRSGDEDAE DAELEASRSR 180
181 LFEQLDSMKL DDGEAEAEED SPPRPAIRRR RRLMRKPMPE PEAASASEEE EVVIAPPPPP 240
241 PYNNETVYLG EAPPPYNNET VTFGATEQDE SLQQQPSPLR VRSLRFTAST STPKSGSKIA 300
301 KRGKKHPEPV ASWMSEQRWA GEPEVMCTLQ HKSIAQEAYK NYTITTSAVC KLVRQLQQQA 360
361 LSLQVHFERS ERVLSGLQAS SLPEALAGAT QLLSHLDDFT ATLERRGVFF NDAKIERRRY 420
421 EQHLEQIRTV SKDTRYSLER QHYINLESLL DDVQLLKRHT LITLRLIFER LVRVLVISIE 480
481 QSQCDLLLRA NINMVATLMN IDYDGFASLS DAFVQNEAVR TLLVVVLDHK QSSVRALALR 540
541 ALATLCCAPQ AINQLGSCGG IEIVRDILQV ESAGERGAIE RREAVSLLAQ ITAAWHGSEH 600
601 RVPGLRDCAE SLVAGLAALL QPECCTQTLL LCAAALNNLS RMEATSHYSI MSNEAIFRLI 660
661 DTLEHQTCGT SVFLYEQIVG MLHNMSLNKK CHSHLANGVI INFITSVYQT EFYQTYGSRA 720
721 ESDAQRRTIK TILHTLTRLA SDSPTLGAEL LEQWHLPDLL RQSLALKPAN SQQQQHLDSS 780
781 YGGDISQLAR QLLRAHGQEQ QLLQATPAPG GSFSSGHQTP EAQTQTNSSG MSSSGSKTKA 840
841 KSSASPLLKF NLTRQESFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
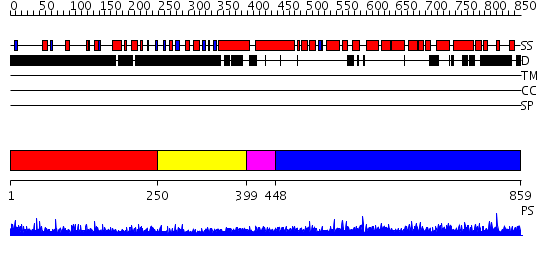
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..249] | 4.236996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [250..398] | 1.214991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [399..447] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [448..859] | 2.05 | No description for 2gl7A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)