













| General Information: |
|
| Name(s) found: |
FBpp0309241
[FlyBase]
sub-PA / FBpp0086041 [FlyBase] |
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 628 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spindle microtubule
[IDA]
spindle midzone [IDA] kinesin complex [ISS] plus-end kinesin complex [ISS] |
| Biological Process: |
spindle midzone assembly involved in meiosis
[IDA]
mitosis [IMP] female meiosis chromosome segregation [IGI][IMP] microtubule-based movement [IEA][ISS] meiotic spindle organization [IGI][IMP] cell cycle [IMP] |
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [IEA][ISS] motor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSKEVSEVP RREVRSFLMA RDPSIDRRFR PRPNKKMRLF DNIQESEEES FSEYSDTESE 60
61 YKYQSSEATE GASCATSAAD SSNVETGPQV FLRLRPVKDA SKAYIVSEEA NVLITSCKVD 120
121 STSNNVNRME KHFGFTSIFD STVGQRDIYD TCVGPKIMEE ECVTIMTYGT SGSGKTYTLL 180
181 GDDVRAGIIP RALENIFTIY QDTVFRSPKL KLINGSIVFL QDDASLKELQ IRKKLLDLCP 240
241 DISAHHQRLK QVIDGDHMFE TKASTDVSVL VWVSFVEIYN ELVYDLLAIP PKQDKLGEVP 300
301 RKNLKIVGNK GHVFIKGLTS VFVTSSEEAL RLLRLGQQRS TYASTSVNAN SSRSHCVFTV 360
361 DILKYNRSGI TTQSSYKFCD LAGSERVNNT GTSGLRLKEA KNINTSLMVL GRCLDAASTV 420
421 QKKKNADIIP YRDSKLTMLL QAALLGKEKL AMIVTVTPLD KYYEENLNVL NFASIAKNII 480
481 FKEPVIKQHR VSYCGFMEFS KMSTCEGGDY TKELEDENVR LQLEIEQLKY DHVLQMQLLE 540
541 EKLRRELTAT YQEIIQNNKK QYEDECEKKL LIAQRESEFM LSSQRRRYEE QIEDLKDEIE 600
601 ELKNPASDTD ISDDPNESKS PIEILDDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
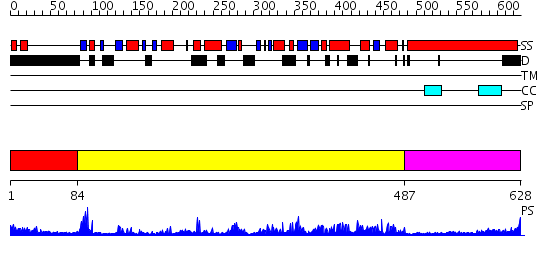
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..486] | 85.522879 | Kif1a head-microtubule complex | |
| 3 | View Details | [487..628] | 5.09691 | No description for 2dfsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)