













| General Information: |
|
| Name(s) found: |
ssp4-PA /
FBpp0086104
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1517 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVETQEIRQ ARQRASVKWL LSKAFNNRVP DNLKEPFYRD HENQERLKPQ IIVELGNATL 60
61 YCQTLANLYS DPNYQSMNHW SIIQTLARKG VPVAESADMP ITETVLIQTN PLRINAHMSV 120
121 IESLMVLYAK EISSGDRVMA AIRRISGNNY QAPTGQSYEQ ALLGWISHAC AALKKRIIKE 180
181 VDAGLPDDNG SRLQTPDIPP VRDFQDLCDG ICLALLISYY CPKVVPWTSV RINYLPAVED 240
241 SIHNILLVCN FSQKHLPYTV MHMTPEDVTY MRGSMKLNLV VLLTDLFNLF EIHPAKCVCY 300
301 PGMDGQVPHS NSFGGGLNRR STPPNEYQTV QSNNFDGNHA EAFVVHKSRG ITTLASMHSQ 360
361 QQQQLHQQQQ HQQQYHQQPL QQHPSQSQLQ IQQQEPLVPA RLRQAKEKTN VESKADERGD 420
421 FVAAGRPSNW EQSRRPSFAG RRSRRNSSSE DSQLTIENFG GSQDQLNTLG RYERDRERKL 480
481 SNTSVGSYPV EPAVAVRSSI ADARGTLQLG YDTDSGSEKQ DRETEKYSMR RQVSVDNVPT 540
541 VSSHNLSNAG SPLPVARHKQ HSSDKDYSSN SGMTPDAYND SRSTSGYDPE STPVRKSSTS 600
601 SMPASPAAWQ LDVGDDDMRS LENASKLSTI RMKLEEKRRR IEQDKRKIEM ALLRHQEKED 660
661 LESCPDVMKW ETMSNESKRT PDMDPVDLDK YQAYNAPVSA YSSRPPSRDP YQQQLHHQQQ 720
721 QPMPMPQPMQ YVNEHGQYMS PPQPAHYMPQ QAQQPQSIYS DNGAAYNHSN HSPYGGTPQY 780
781 RSSVVYDDYG QPTNHFYLHE SSPQPQAHQH PQRRTWAHSA AAAAYEQQQQ IQPSLVDVNA 840
841 WQTQQHQKQK QTWMNRPPSS AGAPSPGSFM LHQNGGGGGG GGGGGELQHL FQVQASPQHG 900
901 QRQVSGSNGV QRQQSLTNLR DNRSPKAPQN MGMPMGMPMQ QEDMMAPQSI CFIGDEEDVD 960
961 ELERNIIESM QSTHISDFVH QQQQQHQHQQ QLQQQQRLQG HSGRGSSSED YDSGEMISNK1020
1021 LNITSGNLTY RIPSPSRPSI QANSFQDPRA MAAASGGEDQ PPEKGFYISF DDEQPKRPKP1080
1081 PLRAKRSPKK ESPPGSRDSV DNQATLKRES LSHLHNNNNI GFGNDDVNSK PVTRHSIHGL1140
1141 NNSNSVKSPG NATYNKYTDE PPIQLRQLAV SGAMSPTSNE RHHLDDVSNQ SPQQTQQPMS1200
1201 PTRLQQSSNN AEAAKNKALV IGADSTNLDP ESVDEMERRK EKIMLLSLQR RQQQEEAKAR1260
1261 KEIEASQKRE KEREKEEERA RKKEEQMARR AAILEQHRLK KAIEEAEREG KTLDRPDLHV1320
1321 KLQSHSSTST TPRLRQQRTT RPRPKTIHVD DASVDISEAS SISSRGKKGS SSNLTGPKLY1380
1381 KQPAAKSNRG IILNAVEYCV FPGVVNREAK QKVLEKIARS EAKHFLVLFR DAGCQFRALY1440
1441 SYQPETDQVT KLYGTGPSQV EEVMFDKFFK YNSGGKCFSQ VHTKHLTVTI DAFTIHNSLW1500
1501 QGKRVQLPSK KDMALVI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
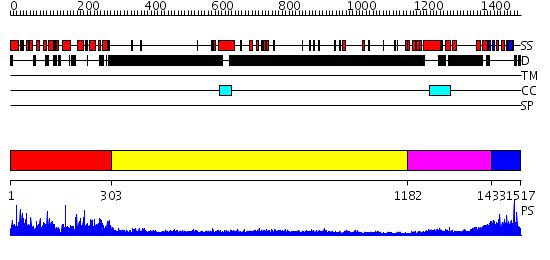
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..302] | 2.26 | Crystal structure of actin-binding domain of mouse plectin | |
| 2 | View Details | [303..1181] | 3.522879 | No description for 1y0fA was found. | |
| 3 | View Details | [1182..1432] | 8.39794 | Heavy meromyosin subfragment | |
| 4 | View Details | [1433..1517] | 64.09691 | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)