













| General Information: |
|
| Name(s) found: |
FBpp0297360
[FlyBase]
FBpp0297359 [FlyBase] CG5065-PC / FBpp0291416 [FlyBase] CG5065-PB / FBpp0291415 [FlyBase] CG5065-PA / FBpp0086254 [FlyBase] |
| Description(s) found:
Found 45 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 625 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
|
| Molecular Function: |
catalytic activity
[IEA]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHAVANKTE TEAAPNSSLK QSAAPQPANS HDAKLLNGTL ARTNGLTHAA SVATSSSGSY 60
61 GSSSAAGSNA GSGGPTSSAS LSIAAGVASS TALPLPPSSN GLQMPYERFR ADDTSYVPIA 120
121 QFYAGRSVFI TGGTGFMGKV LVEKLLRSCP EIRNIYLLIR PKRGQEVSAR LTELLNAPLF 180
181 ESLRQEKPKE LSKVIPISGD ITSEELGISE KDQNLLCRNV SVVFHSAATV KFDEKLKLSV 240
241 TINMLGTKRL VELCHRMLSL DALIHVSTAY CNCDRTDVSE VIYAPPYNPD DIISLINWLP 300
301 EDILDQLTPR LIGKRPNTYT FTKALAEHML LKEAGNLPVA IVRPSIVTAS LNEPFAGWVD 360
361 NFNGPTGLVS ALAKGMFRTM MCEKNYVADM VPVDIVINLM IAAAWRTATR KSNNLLIYNC 420
421 CTGQRNPIIW SEFVKHAMTS VRKHPLEGCL WYPTGDLRMN RPMNTLNCIA KHFLPAYILD 480
481 GVARIMGKKP FVVNVQNKIA KAVECLEYFA TRQWRFKDDN VHALLHTLSP KDREIFVFDV 540
541 RHINWDKYVE RYVLGFREFL FKQRPESLPA SRKRMLRLYY LHQLTKLVAV LLTWRFLMSR 600
601 SKRLNDLWSS FLENALRMAR LIPFL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
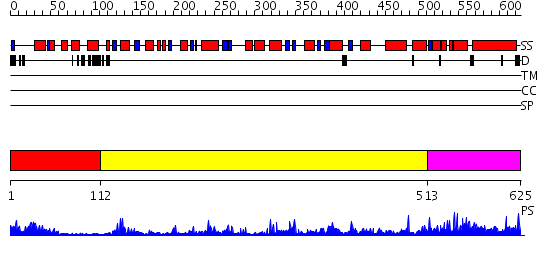
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 8.09691 | No description for 2gdwA was found. | |
| 2 | View Details | [112..512] | 64.30103 | dTDP-glucose 4,6-dehydratase (RmlB) | |
| 3 | View Details | [513..625] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.42 |
Source: Reynolds et al. (2008)