













| General Information: |
|
| Name(s) found: |
csul-PA /
FBpp0088810
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 610 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA][NAS]
|
| Biological Process: |
pole plasm protein localization
[IMP]
intracellular mRNA localization [IMP] P granule organization [IMP] mitosis [NAS] pole plasm assembly [IMP][NAS] |
| Molecular Function: |
protein methyltransferase activity
[IMP][NAS]
methyltransferase activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYYVCLHQE GVNSIPKLIE KAFANNYNVV STSINANMLP FEPHESDPTY PATILSGSDW 60
61 NSKVIFTMSD VNVDSPNDKL REHAKEVFMR DVAWAEHLQN VGNLMVRLRG PENENLASIV 120
121 LAKTKDDFPS GNWFIQVPIT NPELATFEHR KDATAEEVAE AESNDPWNWW NNLRMVTKHS 180
181 TKVKVVIELN DADRPSKETV RRWLGEPIEA IIIPSSLFVR NRSNYCVLKK EWQLIVGHFI 240
241 SVRANIIIST NPNDKALCQY ADYVNKLIND NCDKHMLNSY ENMLEIPLQP LCDNLDTYTY 300
301 EVFETDPVKY KLYQDAVQAA LLDRVSAAEA KTKLTVVMLL GGGRGPLARA VFNAAELTKR 360
361 KVRLYIIEKN PNAIRTLSNM VKTLWADKDV HIFSKDMRDF SPPELADIMV SELLGSFGDN 420
421 ELSPECLDGA LKLLKPDGIS IPYKSTSYIN PLMSAVLHQN VCQLLPTYPA FDYGYVSLLK 480
481 NIYHIDEPQA LFEFVHPNRA ENIDNTRCKT VSFKVNKDCV LHGIGGYFDT HLYKDICLSI 540
541 NPLTHTPGMF SWFPMFFATR PRTLREGQTI SIQFWRCVDA TKVWYEWQVV NSPDDWEHHN 600
601 TRGTGYNMRL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
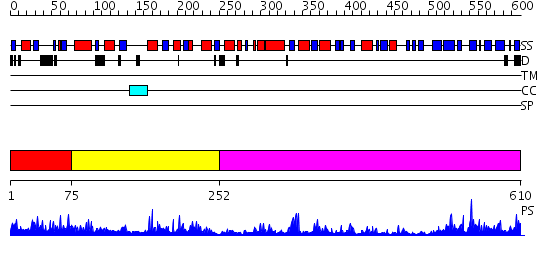
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..251] | 1.476979 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [252..610] | 55.39794 | Protein arginine N-methyltransferase 1, PRMT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)