













| General Information: |
|
| Name(s) found: |
CG8079-PB /
FBpp0086499
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 599 amino acids |
Gene Ontology: |
|
| Cellular Component: |
precatalytic spliceosome
[IDA]
|
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IC]
|
| Molecular Function: |
nucleic acid binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLPAEEEDL ADTVAKYDSI ELKSIQDLET CEQQQLLQYV EKLHGIIRKY DTKLASYKQK 60
61 LLHYVSREEK VSCDKASQAT ESELQECQYS MENDEDTNKK DQPNDLSATD AFSFVDEMRQ 120
121 AAKHAENLNN FVYEHTSGMY YDPKTGYYYN AEYGLYYDGN NGCYYSYDHA KDSYEFHSQA 180
181 QANDAAKPES EDEDLEVQFD ELGGVITDHE TLKKIKAEKQ KAKDQAEKSK RKAKKKKSKK 240
241 HSKKRSKKER RHKSKKRHRH SDDERSNDAE EGELSQSSDS SSDSSNEDSS SNTEDSSVPV 300
301 FKAAGRFQDI AKKYPPSLRI IVQETNVESL KVGSLHLITY KGGSLGREGA HDVIIPDVNV 360
361 SKCHLKFKYE NKLGIYQCLD LGSRNGTILN GSPMSSDAMD LVHGSVITLG QTRLLCHVHE 420
421 GNSTCGLCEP GLLIENSPPV VAAVASSTAS VLSHKEQLKK LQRKYGLENE KFVDTSGNGQ 480
481 SNYNDRAATR RVQVGSSTDK EKTEVACVNT EIGSSNKGFK MLSKLGWQKG EKLGKTNASA 540
541 GLLEPINVVA NEGTSGLGNS DPVLSSSRTI DKRKLANLKI TQARYQRASD MFGQSDDSD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
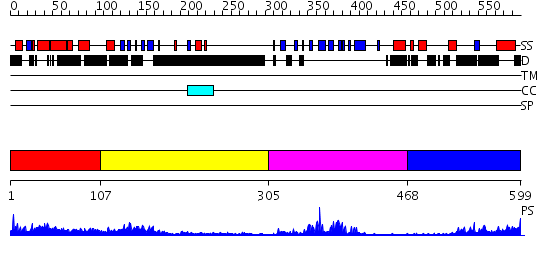
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..304] | 5.250993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [305..467] | 14.0 | Solution Structure of human Ki67 FHA Domain | |
| 4 | View Details | [468..599] | 18.256996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)