













| General Information: |
|
| Name(s) found: |
Rcd1-PA /
FBpp0086708
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1001 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
centriole replication
[IMP]
|
| Molecular Function: |
hydrolase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSFFDCFND FLQSGGNLVQ NVSQEQEQKI ADQELDGVIE LMPPTTPTPS ESSSTISAPS 60
61 ESSFSEKMEH SYMRDTPVRS DVHNGLTPAR NILVRHPPQC PSCHTYPPQH EELSEIQQAH 120
121 VPPYDEVVAK EAMNECARIA KYVKNNNSDE QDWVARVNKF NWTPMQEMVF EKVCSILDQD 180
181 QLARLANDKR QHEPIHRRIS SDKSASRFRK LLASVAWESR ITQWIHALLM EHLSPSYMAS 240
241 YLEILQTLKT KLPTLVDKML FGRPLNNSQE LLAPVMKKRW EPNIMPKSRQ LTHNAIMVVL 300
301 PNMPTTGTVS DRMQKWYQAL STITQVVQIS LPNTNNRIGN QNLDHVAETI VSITRVKINE 360
361 LRTENPSRGI ILVGFNAGAA LALQVALSES VACVVCMGFA YNTIRGPRGT PDDRMLDIKA 420
421 PILFVIGQNS ARTSQEEMEG LRERMQSESS LVVVGSADDA LRVPKSKRRI EGVTQSMVDY 480
481 MVVEEIFEFV NRTLSNPPGP RMPTCLMHQQ GYQRLPKQSP HILADGNANK AVQQMRKRKM 540
541 DGGLDDSMGQ PSKTKFVPHN DAQDYGDAQT TAGGSASLPK VITPGVLGKQ LPPVNLAAGT 600
601 KIKMIPSSQF VQIKSLPSQS KLINYTLNKP SVGTSSTANI GSVVKTLPCS SVSGQQIITL 660
661 KTPSGQTQQF ATAATSTGAA GSSTSTGTGQ KKYTVFKNAN GMTMLHLTKS VPTTGSSSGS 720
721 ANMDLSNIID MPIVFADNEG NIPEHQQADK DIKPAPIRSA NKSPLIISQK IIKEANKPSG 780
781 MVTSGSITAV SKPGNIVLNK GLQQLLNSSP GGTLATQNKV VYLNRSTIKP MGNVAPGTHR 840
841 VPIRIVSTPN TGGAVTTTAS SSPIMVDPAT GSKVAKTVNV QTIKPTGSVL AQGTIRQAGN 900
901 VGTKSYLQMI NTAPGASKTV AGDGKGSVRN IYFKSATGLK QLPVQMLGNR IPGGAVISST 960
961 ASASSGSPGL RRVVNIGPVS KVTATSTTSS SIAATLTHNK M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
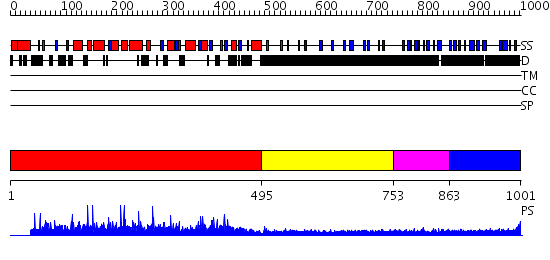
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..494] | 0.999 | Epoxide hydrolase, N-terminal domain; Mammalian epoxide hydrolase, C-terminal domain | |
| 2 | View Details | [495..752] | 1.239994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [753..862] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [863..1001] | 4.249997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)