













| General Information: |
|
| Name(s) found: |
FBpp0292098
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 716 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transport vesicle
[ISS]
|
| Biological Process: |
locomotory behavior
[IMP]
glutamine metabolic process [IEA] memory [IMP] |
| Molecular Function: |
carbon-monoxide oxygenase activity
[ISS]
glutaminase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKGVPTLAG RNLRLLTTQA QVLVHQRAKA SAAEPVEDFD QETDLLKRAK FGHTGKQLQQ 60
61 KIINEKNLRA PPLMNLVFAN RKLSFYDTSS TPKTKEDNAK LSQRRHYSMR PHREQEQRNA 120
121 EDVLFDMFAS EETGLISMGK FLAGLKTTGI RRNDPRVREL MDNLKKVHKL NNYETGSSAE 180
181 TQHLNRETFK AVVAPNIVLI AKAFRQQFVI PDFTSFVKDI EDIYNRCKTN TLGKLADYIP 240
241 QLARYNPDSW GLSICTIDGQ RFSIGDVDVP FTLQSCSKPL TYAIALEKLG PKVVHSYVGQ 300
301 EPSGRNFNEL VLDQNKKPHN PMINAGAILT CSLMNALVKP DMTSAEIFDY TMSWFKRLSG 360
361 GEYIGFNNAV FLSEREAADR NYALGFYMRE NKCFPKRTNL KEVMDFYFQC CSMETNCEAM 420
421 SVIAASLANG GICPTTEEKV FRPEVIRDVL SIMHSCGTYD YSGQFAFKVG LPAKSGVSGG 480
481 MMLVIPNVMG IFAWSPPLDH LGNTVRGLQF CEELVSMFNF HRYDNLKHLS NKKDPRKHRY 540
541 ETKGLSIVNL LFSAASGDVT ALRRHRLSGM DITLADYDGR TALHLAASEG HLECVKFLLE 600
601 QCHVPHNPKD RWGNLPVDEA ENFGHCHVVE FLRSWAEKAD QSSEECKPEA VTSKTQADEE 660
661 ICSTSDLETS PTTSPIPTPV ESGSRAGSGR SSPVPSDAGS TASAGSSIDD KTKPGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
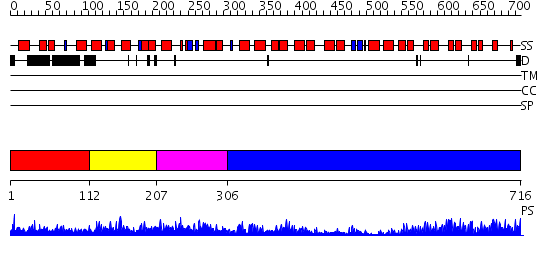
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [112..206] | 1.05 | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | |
| 3 | View Details | [207..305] | 49.522879 | No description for 2dfwA was found. | |
| 4 | View Details | [306..716] | 69.522879 | Ankyrin-R |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)