













| General Information: |
|
| Name(s) found: |
CG8841-PB /
FBpp0087074
[FlyBase]
CG8841-PA / FBpp0087073 [FlyBase] CG8841-PC / FBpp0087072 [FlyBase] |
| Description(s) found:
Found 43 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 837 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNTDSKLNF RKAIVQLTQK NQKIDPSDEQ FWEQFWQGHQ TTLEDVFALV TSSEIRQIRN 60
61 ENPANLATLC YKAVEKLAQA VDSSCRTQAE QQCVLNCVRL LVRCLPYIFE DEKWRDFFWS 120
121 SLPSQDKTMP LAQSLLNATC DLLFCPDFTV TATRRAGPEK AEELANIDSC EYIWEAGVGF 180
181 AQSPPHNAHM ERRRTELLKL LLTCFSEPMY RSPQQSEEPN KWIAYFTSAD NRHALPLFTS 240
241 FLNTVCSYDP VGFGVPYNHL LFADTTEPLV EACLQLLIVT LDHDMVVQQQ LTQPGQASYD 300
301 EGNCGDNLFI NYLSRVHRDE DFHFVLKGIT RLLNNPLVQN YLPNSTKRLH CHQELLILFW 360
361 KICDYNKKFL YFVLKSSDVL DILIPILYHL NYSRADQSRV GLMHIGVFIL LLLSGERNFG 420
421 VRLNKAYSAT VPMDIPVFTG THADLLITVF HKIIATGHQR LQPLFDCLLT ILVNVSPYLK 480
481 TLSMVASVKM LHLLEAFSTP WFLLSAPSNH HLVFFLLEIF NNIIQYQFDG NSNLVYTIIR 540
541 KRHVFHAMAN LPTDMAGIAK CLSGRKTGGK FNLPRVPQRR TPAVSQELPS AHVPEEYNEE 600
601 DEDEDEEEII NEEPKAEEDL ESETESHERS QAGELQSDVL TAQPAEPGTL KTSLLDTPGI 660
661 TQMTEREQAH PNDKPQVEDS TDIVPYDRSA ASTPTDERKS TSPTELSRLS VAHRASIRMV 720
721 PGESDRWTPT PEWIVSWRSK LPLQTIMRLL QVLVPQVEKI CIDKGLTDES EILKFLQHGT 780
781 LVGLLPVPHP ILIRKYQANA GTTAWFRTYI WGVIYLRNVE PAIWYDTEVK LFEIQRV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
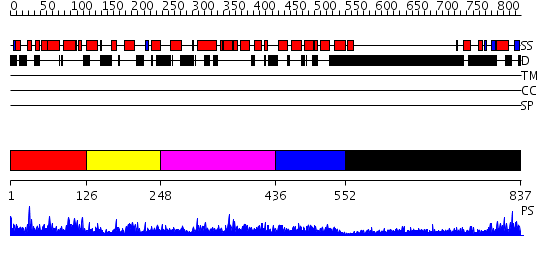
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 1.043954 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [126..247] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [248..435] | 1.048957 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [436..551] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [552..837] | 1.49 | No description for 2grxC was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)