













| General Information: |
|
| Name(s) found: |
gi|24652774
[NCBI NR]
gi|21627440 [NCBI NR] gi|15292405 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
chromatin remodeling
[NAS]
nervous system development [IMP] |
| Molecular Function: |
DNA binding
[IEA]
zinc ion binding [IEA] protein binding [IEA] chromatin binding [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRERIIDLE ANIERRYLKP PLGSQTGDAH LAVIAQNQHT TTQTQNSASA AAYLLQMQQQ 60
61 QQQQQLAQQQ QQQQQGSGAG NSLNPSSFNE RTMALAAAAA ASGPGNATGV ANSAVVAGAT 120
121 PCESGSGEPN SGNASPASNC DSDRDEKVEQ IPKGLVQWRD AVSRSHTTAQ LAMALYVLES 180
181 CVAWDKSIMK ANCQFCTSGE NEDKLLLCDG CDKGYHTYCF KPKMDNIPDG DWYCYECVNK 240
241 ATNERKCIVC GGHRPSPVGK MIYCDLCPRA YHADCYIPPL LKVPRGKWYC HGCISRAPPP 300
301 KKRSAGGTSG SSSKSRRDRD RESGGSAKRR SDNSKTPAME HMQQQQMPLA GGDSHHHHHQ 360
361 QPPSLNSSHD ESMNSLPAAP LSPAHSVVSA TNYDDQHHAN NSVDGSSRFH AHLIPPSNNG 420
421 TAALLEDVPG GANVMPGVYP VYTPVAAGNF SAGLINQAPV QPAMPFANVV AMSPRAVTPT 480
481 RTRTPTPTPA PTPPPPPPTP LLMQASPTAT ALHVNACQSP PQQQHAQLMT MPSPPAIGVG 540
541 TATNQMSPPP INIHAIQEAK EKLKQEKKEK HATKKLMKEL AVCKTLLGEM ELHEDSWPFL 600
601 LPVNTKQFPT YRKIIKTPMD LSTIKKKLQD LSYKTREDFC VDVRQIFDNC EMFNEDDSPV 660
661 GKAGHGMRKF FESRWGELTD KHS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
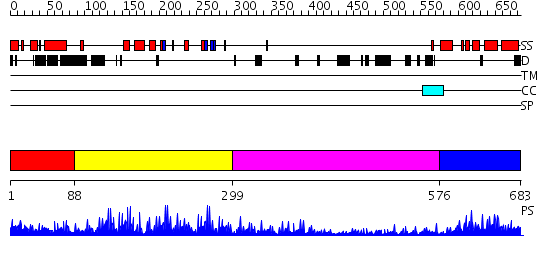
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..298] | 36.0 | No description for 2i13A was found. | |
| 3 | View Details | [299..575] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [576..683] | 25.221849 | No description for 2e7oA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)