













| General Information: |
|
| Name(s) found: |
skf-PB /
FBpp0113061
[FlyBase]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 556 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[ISS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein binding
[IEA]
guanylate kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLEAVMASDI NWDPALSKLI STLKESENLS NDQEIDFLKA LLESKELNAL VNVHTKVAKV 60
61 GRDDRLAPVL STSAQVLYEV LEQLSQHCHL NDDCKEAFHL LQDSHLQHLL FAHDAIAQKD 120
121 FYPHLPEAPV EMDEDEETIK IVQLVKSNEP LTGAQSTEPI VGATIKTDEE SGKIIIARIM 180
181 HGGAADRSGL IHVGDEVIEV NNINVEGKTP GDVLTILQNS EGTITFKLVP ADNKGAQRES 240
241 KVRVRAHFDY NPDVDPYIPC KEAGLAFQRG DVLHIVAQDD AYWWQARKEH ERSARAGLIP 300
301 SRALQERRIL HDRTQKNGTD LDSKQNDDFD REQIATYEEV AKLYPRPGVF RPIVLIGAPG 360
361 VGRNELRRRL IARDPEKFRS PVPYTTRPMR TGEVAGREYI FVAREKMDAD IEAGKFVEHG 420
421 EYKGHLYGTS AESVKSIVNA GCVCVLSPHY QAIKTLRTAQ LKPFLIHVKP PELDILKATR 480
481 TEARAKSTFD EANARSFTDE EFEDMIKSAE RIDFLYGHFF DVELVNGELV NAFEQLVQNV 540
541 QRLENEPVWA PSMWVQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
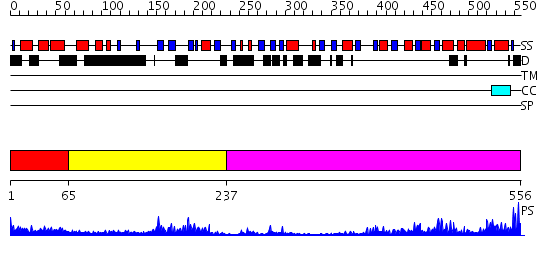
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | 3.30103 | 2.1 Angstrom crystal structure of the PALS-1-L27N and PATJ L27 heterodimer complex | |
| 2 | View Details | [65..236] | 24.39794 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 3 | View Details | [237..556] | 45.09691 | Psd-95; Guanylate kinase-like domain of Psd-95 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)