













| General Information: |
|
| Name(s) found: |
CAP-PA /
FBpp0087430
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 2376 amino acids |
Gene Ontology: |
|
| Cellular Component: |
focal adhesion
[ISS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
vinculin binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNNRNRNRN RNRNKRNRNQ NQNQNPSQNQ NENHQQQAEG AEDREEQQDF QTQIAVAQSS 60
61 SFVDDNGNSG SVSNGPTENS EEVTNTLEKT EKKEEIFEQP ATVIQKEADS DAEMGAKNSK 120
121 HRKEKSDKQA SNGKSEEQVR PPPTIIRRPP GSPRQAKVIV HRIVREEDTA NGKAQSPEPP 180
181 KAVESKEQHM EVEDSKKEKP EAQQEIETKE ERENEPSPKE LPESSHLSED AQQKHVEVEE 240
241 NQQVYDEVDY SKDETTDNNP QQEQHDSKEP EQYVLQLEKA MEFVENAHQQ HVAAVQSYQK 300
301 QRDTEQNAQQ QEQNEAQHLQ ESTKAHQQNI AVPQILVDPK KREEGLAQQL HHDVKEEPIL 360
361 NNAQQQNDDT APKSPKEVTI KVQFQTEDPY SQQPPPQKPT SKVIIHQIHV ETTDEEQNAK 420
421 PTFEEVSSTT GSLAGTLSPP PRYLVESPKN VSSGQFSNFS RNVQIQEMEL NSDFSSGEFN 480
481 SLQSPLVCEV DSESEGSVVL GPERSPSDQQ VPSSSRVEQH EQVRQKRAQY RNALESHFMP 540
541 QLLNPRYLDS ILEENEWRNS TASSGGSDQT GIRTPKLNET FPRSQLDFSR RHKRREEAPS 600
601 PLKLETKLLE ESTDLESCTR LQSALSPQSE DAELVYLSSS ASSSVSDLME LELEQAAALA 660
661 ERALVDLDTD ASRLIARPDD EMSSTSTTTA DRSETEAETE TETEREGEQS RESTPVNANP 720
721 TLASSQSSLL SAATPTPTPT PTPADREQLA KDTNVSGGST VSFGAASPLA ATREEFVRNM 780
781 DKVRELIEMT RREQEQGELP RSPSPPPVPP PPASVPPYSP ESSSFHLASL QLKRQESNDS 840
841 HCSDSTTHSQ CTAINLASPP PPPTAQPPTP PLRQKPAPPP VPQPVSPPAP PTPQSEPELS 900
901 VADSVANANA DVDADADTDT EAIKKLRLLC TEQLASMPYG EQVLEELASV AQNISDQSQN 960
961 KMPYPMPQLP HIKELQLNAN ESKSSSAWLG LPTQSDPKLL VCLSPGQRDL VNNQTQPDDL1020
1021 LDAHQKFVER RGYHELSKAQ VLEQDHQQQQ SEMLKTAAMM RELRKSLSPP APPVPPPPVP1080
1081 VKSAETAAKA TAHENAKRDD ASEQKQKIAA CESSSLENKP RRAADLGAQQ SAEQRQSSST1140
1141 TTSSHKATTE TMSSDTAKFP TLDSMESELA RMFPQHKGDI FEEQRKRFSN IEFPSHQPVS1200
1201 QTKRYSNIET SSYESKKRME NGQVVYDVST SSHEKKEQGD PPKDQPVPPV PPPPIMSATK1260
1261 LNGNTFIDGD VAPKNSQPSR ESGSGSGNGT YEEFRQRAKA AADAFGEQRE QQNGLDQDRV1320
1321 FKDFDRLSQQ MHAELQSTRE KREKSASMYD LSGFTRPASG HPRLDELQQR RHAHMQELER1380
1381 EIERSAKSRQ ERMSSVPRQM EATPPRTHEI PIELEPRSRR AESLCNLNEP PPRPHTTVGH1440
1441 YNHPVVQDDW SRYANDLGYS ENIARPFARE VEICYQRQNQ RTPHCIRAPR LSASTNDLSS1500
1501 SSQYSYDTFN AYGGRRTHAP MLNQAQQQQR PHYGSCYSMI ERDPNPRYIS TTSRRGVSPA1560
1561 PPPVATPQQQ QVPPPAYDRQ QRRSSLPREL HEQQLKYILS KEEELKFEVE RLQQERRRLM1620
1621 EEMQRAPVLP APQRRESYRP AAKLPTLSED EVFRQQMAEE WMNKVAEREE RRQHKIIRIS1680
1681 KIEDEHDHSA VDKATISDEF LDRVKERRHK LSMPADSDWE SGAESQPQPA AQSQPESDVE1740
1741 APPVRILEGQ AEANLRQLPR HLREFAKFST SEQLPDGAQM ERHEEQERRE EATDNAHSSA1800
1801 TKKTSIVKTY KVSRLPPSVQ ARPSYKSNGY VSEPEPNYDS DYSTVRYRTQ NPHRVQSVSS1860
1861 AVNVRNLNQD EKLYGTMPNP IKSAQNSYKN QPGRIENYTT GHSSVSEKEK KEWWDEVMDI1920
1921 FNGNLEQSKL SPLYTEGNLS RALAKESGYT SDSNLVFRKK EVPVSSPLSP VEQKQAYKSL1980
1981 QAGGEPPLLG FRKPAPEKPR DLDPNAPPIP PQPPVKGLSS YDFPYSTDTV DGSDVNIHFK2040
2041 TPIRHEQRQN LSEEELAIRQ AEHMQKLYHE ERRRKYLQEL QDMNSRRHTD NFTPSQKSPI2100
2101 ALNRYDDFPT DVTLKSLVGP KTVARALFNF QGQTSKELSF RKGDTIYIRR QIDANWYEGE2160
2161 HNAMIGLLPA SYVEIVSRDG ARTPSKRPSE GQARAKYNFQ AQSGIELSLN KGELVTLTRR2220
2221 VDGNWFEGKI ANRKGIFPCS YVEVLTDIGA EDIAARTTTV ITSQSTTNLR PNLDVLRTNI2280
2281 NNEFNTLTQN GAQPPNGILK ETRTLHKTDA LHVDTSSEPL AYRALYKYRP QNSDELELLE2340
2341 GDVVHVLEKC DDGWFVGTSQ RTGCFGTFPG NYVERA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
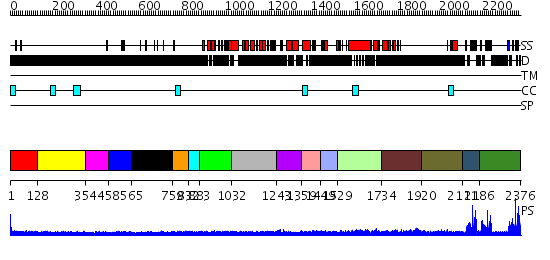
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [128..353] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [354..457] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [458..564] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [565..758] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [759..831] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [832..882] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [883..1031] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1032..1242] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1243..1358] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1359..1448] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1449..1528] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1529..1733] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1734..1919] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [1920..2110] | N/A | No confident structure predictions are available. | |
| 16 | View Details | [2111..2185] | 24.522879 | p47pox (neutrophil cytosolic factor 1) | |
| 17 | View Details | [2186..2376] | 32.69897 | Growth factor receptor-bound protein 2 (GRB2), N- and C-terminal domains; Growth factor receptor-bound protein 2 (GRB2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. | |||||||||||||||
| 7 | No functions predicted. | |||||||||||||||
| 8 | No functions predicted. | |||||||||||||||
| 9 | No functions predicted. | |||||||||||||||
| 10 | No functions predicted. | |||||||||||||||
| 11 | No functions predicted. | |||||||||||||||
| 12 | No functions predicted. | |||||||||||||||
| 13 | No functions predicted. | |||||||||||||||
| 14 | No functions predicted. | |||||||||||||||
| 15 | No functions predicted. | |||||||||||||||
| 16 | No functions predicted. | |||||||||||||||
| 17 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)