













| General Information: |
|
| Name(s) found: |
FBpp0311591
[FlyBase]
FBpp0304256 [FlyBase] Hdc-PA / FBpp0087422 [FlyBase] |
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 847 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
compound eye photoreceptor development
[NAS]
cellular amino acid and derivative metabolic process [IEA] carboxylic acid metabolic process [IEA] |
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
histidine decarboxylase activity [IMP][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFKEYRQRG KEMVDYIADY LENIRERRVF PDVSPGYMRQ LLPESAPIEG EPWPKIFSDV 60
61 ERIVMPGITH WQSPHMHAYF PALNSMPSLL GDMLADAINC LGFTWASSPA CTELEIIVMN 120
121 WLGKMIGLPD AFLHLSSQSQ GGGVLQTTAS EATLVCLLAG RTRAIQRFHE RHPGYQDAEI 180
181 NARLVAYCSD QAHSSVEKAA LIGLVRMRYI EADDDLAMRG KLLREAIEDD IKQGLVPFWV 240
241 CATLGTTGSC SFDNLEEIGI VCAEHHLWLH VDAAYAGSAF ICPEFRTWLR GIERADSIAF 300
301 NPSKWLMVHF DATALWVRDS TAVHRTFNVE PLYLQHENSG VAVDFMHWQI PLSRRFRALK 360
361 VWFVLRSYGI KGLQRHIREG VRLAQKFEAL VLADHRFELP AKRHLGLVVF RIRGDNEITE 420
421 KLLKRLNHRG NLHCIPSSLK GQYVIRFTIT STHTTLDDIV KDWMEIRQVA STVLEEMNIT 480
481 ISNRVYLKET KEKNEAFGSS LLLSNSPLSP KVVNGSFAAI FDADEFLAKT YAGVRIAHQE 540
541 SPSMRRRVRG ILMSGKQFSL DSHMDVVVQT TLDAGNGATR TSTTNSYGHT TSAAQANSER 600
601 QASIQEDNEE SPEETELLSL CRTSNVPSPE HAHSLSTPSR SCSSSSHSLI HSLTQSSPRS 660
661 SPVNQFRPIT LCAVPSQSQL SMPLAMPLPN RNVTVSVDSL LNPVTTCNVY HGKRFLEPLE 720
721 NLAQTSASFS SSIFRLPTPI ATPTRESPED PDWPAKTFSQ LLLERYSSQS QSLGNNSSTE 780
781 SSSLSGGATP TPTPMSSLDE LVTPLLLSFA SPSQPMLSAH GIGEGQREQG SDSDATVCST 840
841 TSSMESL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
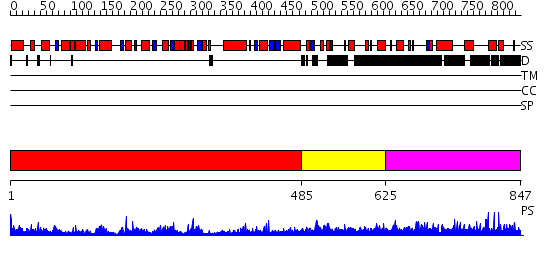
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..484] | 118.0 | DOPA decarboxylase | |
| 2 | View Details | [485..624] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [625..847] | 5.057997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)