













| General Information: |
|
| Name(s) found: |
Pfk-PB /
FBpp0087508
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 788 amino acids |
Gene Ontology: |
|
| Cellular Component: |
6-phosphofructokinase complex
[IC][IEA]
|
| Biological Process: |
glycolysis
[IMP][NAS]
|
| Molecular Function: |
6-phosphofructokinase activity
[IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSEINQRFL ARGSQKDKGL AVFTSGGDSQ GMNAAVRACV RMAIYLGCKV YFIREGYQGM 60
61 VDGGDCIQEA NWASVSSIIH RGGTIIGSAR CQDFRERQGR LKAANNLIQR GITNLVVIGG 120
121 DGSLTGANLF RQEWSSLLDE LVKNKTITTE QQEKFNVLHI VGLVGSIDND FCGTDMTIGT 180
181 DTALHRIIEA IDAISSTAYS HQRTFIMEVM GRHCGYLALV GGLACEADFI FIPEMPPKVD 240
241 WPDRLCSQLA QERSAGQRLN IVIVAEGAMD REGHPITAED VKKVIDERLK HDARITVLGH 300
301 VQRGGNPSAF DRILACRMGA EATLALMEAT KDSVPVVISL DGNQAVRVPL MECVERTQAV 360
361 AKAMAEKRWA DAVKLRGRSF ERNLETYKML TRLKPPKENF DADGKGIEGY RLAVMHIGAP 420
421 ACGMNAAVRS FVRNAIYRGD VVYGINDGVE GLIAGNVREL GWSDVSGWVG QGGAYLGTKR 480
481 TLPEGKFKEI AARLKEFKIQ GLLIIGGFES YHAAGQIADQ RDNYPQFCIP IVVIPSTISN 540
541 NVPGTEFSLG CDTGLNEITE ICDRIRQSAQ GTKRRVFVIE TMGGYCGYLA TLAGLAGGAD 600
601 AAYIYEEKFS IKDLQQDVYH MASKMAEGVS RGLILRNEKA SENYSTDFIY RLYSEEGKGL 660
661 FTCRMNILGH MQQGGSPTPF DRNMGTKMAA KCVDWLAAQI KANIDANGVV NCKSPDTATL 720
721 LGIVSRQYRF SPLVDLIAET NFDQRIPKKQ WWLRLRPLLR ILAKHDSAYE EEGMYITVEE 780
781 ECDTDAVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
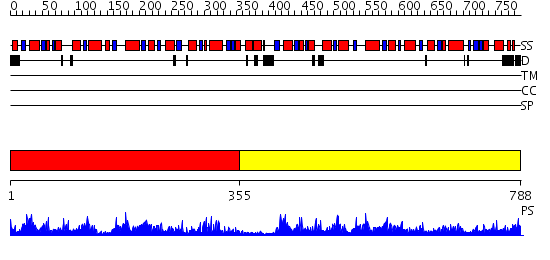
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 105.0 | ATP-dependent phosphofructokinase | |
| 2 | View Details | [355..788] | 79.0 | No description for 2higA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)