













| General Information: |
|
| Name(s) found: |
Rme-8-PA /
FBpp0087699
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 2408 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
border follicle cell migration
[IMP]
endocytosis [NAS] receptor-mediated endocytosis [IMP] |
| Molecular Function: |
heat shock protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLPPKENVDL ECFLVTKHSW KGRYKRILSI GTAGISTYNP DKLDLTNRWS YSDIVAAAPS 60
61 KTTNIPHDFV ITIKKDKKLD TIKLSSEYRN DILSSILKYY KEFADKPKNA QRFSAYKYHW 120
121 SGISLPTVLE VTPCSLDQLD PTTNDVLASY MYKDIEGIIA GISDYEGGIV MAYGGFSRLH 180
181 LFKALNHHEI VQNIAQRSAQ FLGIETKILK SQITLEQFER QRFGKFSGDQ FQTSSTEFSV 240
241 HKITPRHPDP VKRILCLTEV TLLERDPQTY SVCTLRPLTD VFALVRDKDN LQRFSIEYKN 300
301 GIVRNYSTND RDSLLATVLD AVRSSGNQDV HVRIGNTPRG KRYVPLNSSV DEETEANLMR 360
361 LVINNFQNPS KRYEILERFN ANVPHSGLNY SVTQDSLFAE NKDKLILSAL QALAQKELDS 420
421 PTAQLSNLEL EAIFHALTRL LASKVGYAAF TNLPGFREII GTKVVAALRR KDLAVTYSAI 480
481 DMINSLMHSV NSDHDLKQEQ LNKSSILSSK SFLETLLNMW TTHVSHGSGA LVLSAMLDFM 540
541 TFALCVPYSE TTDGKQFDTL LELVADRGRY LYKLFQHPSL AIVKGAGLVL RAIIEEGELQ 600
601 VAQQMQALAL DEAALCRHLL VALYTPSNDP TQTTQRQLSR HLIGLWLTDS EEAMELFKRI 660
661 FPAGLLTFLE SEETVPESDV EEDKLNFRDN LKFAIQHSNK TRKNVIEKHL QGIKHWGMNL 720
721 IEKQDGAAQA LKNRPVVLRN RRQKKKTSDV VVNLPFFFYS FAKDHSLPNL IWNHKTREEL 780
781 RMCLENELRQ FLNDRDLAGQ MIVAWNYQEF EVAYQCLADE IKIGDYYIRL ILEKDDWPQN 840
841 LVKDPIELFN ALYRRVLCRQ RVNDDQMTVF SLQALAKVYR RYHAEIGKFN DMSYILQLSD 900
901 RCLSPSMRDA LINLISCLVL EKSNSRALID HVQCLVDLIT LAHLHKGRAQ LNTKTNVIEA 960
961 GPNMGTYEEK DWYYNIEKDG QKPERQGPIT YSELKELWQK GLITPKTRCW AIGMDGWRSL1020
1021 QQIPQLKWCL IAKGTPLYDE TELSSKILDI LIKCTSFFPS RTQNGVAVLI PGPKLSRKLS1080
1081 EFICLPHVVQ VCLTHDPGLL ERVATLLYQI MEDNPEMPKV YLTGVFYFML MYTGSNILPI1140
1141 TRFLKMTHMK QGFRSEETSQ SGIMHRSILG QLLPEAMVCF LENYSAEKFA EIFLGEFDTP1200
1201 EVIWSSEMRR LLIEKIAAHI ADFTPRLRGH TMARYPYLAI PVISYPQLEN ELFCHIYYLR1260
1261 HLCDTQKFPN WPISDPVQLL KHTLDAWRKE VEKKPPQMTI QQAYQDLGID LTKTPKPDES1320
1321 MIRKSYYKLA QMYHPDKNPN GREIFEKVNQ AYEFLCSRNV WSSGGPDPNN IVLILRTQSI1380
1381 LFERYPDVLR PYKYAGYPQL IKTIRLETRD DELFSKEAQL LTAASELCYH TVHCSALNAE1440
1441 ELRREEGIEA LLEAYTRCVS ILGVDSKPDS LHYQVISNVT RCFEVACNFE KCKQKIIQLP1500
1501 QLLSDVCRVV YFKHTLSVSL VTSLAANNYD LQCQLSRNGV LWSLLLFIFE YDYTLDESGV1560
1561 DVSDKSNQQQ LANNLAKMAV LGCIALAGYS MELRQKPVTG SETNSPAAKA PPVIKPKPSV1620
1621 SAASSSTYTQ NAHNPLQSKQ LAITSGKEKE SDTSSGSSDT ASSTPTESEQ QQQLTKTRTS1680
1681 AIQQKYIVTG EAKNSLIKQV LDRLLTRYIS NQLATVRDSD VLKLLTANTR NPYLIWDNGT1740
1741 RAQLKDFLEQ QRTASAKETH EDIAQVSELV SSFEFDAHKD ELQIGGIFIR IYNDMPTHPI1800
1801 AQPKLFIMDL LEYLKHAYQF LQYKKNPAST AAPVNATPKM GNDGILTPTL APNHPQLQQA1860
1861 STGKSGTTFD EVLTAYNRSK SRKKLETDAL TEQQLALQQS KYDFSSDGKI ELHITMVLKA1920
1921 LIAVIKANAE VEIQCIGNFD MIFGFLASNI FPDNSTVKAV ALEVVSLVSR NKECVSEVAA1980
1981 CEILGNYLVA LKDPELRASQ VKVLETLSGL MNVQEMIKEA QAKGATIYLL DMFCNSRNPQ2040
2041 IREMCAGILA KMTADRLSGP KVRITVSKFL PALFIDAMVE SPATSVQLFE SIHEHPELIW2100
2101 NDTTRSNVCD AVADTCQRFY QLQKANSRHV WKDPEILKDI VSNEIVVAGV YLRLFVSNPA2160
2161 WTLRKPKQFL SDLLDFVVEQ IGKSSSEQDV LDLSTTALVE LLRSQPNLAD DIPVLGHIPK2220
2221 LFNLLSVQPK NTLSVLHQLS LSEFCVSAIS QTECVAPLKK CMEHNRDCIE KACEALSRLF2280
2281 KHQHDSLISQ SLEVGLIPFL LGLLDSRLEF VDNPSAVKAQ IVAALKGMTH NLNYGDRVTQ2340
2341 ILLKHPVWAE FKDQRHDLFI ADTTIRGYLT GVNPTAGYLT AGPAQSVEVL TSPPPIDRDD2400
2401 PSARPPID |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
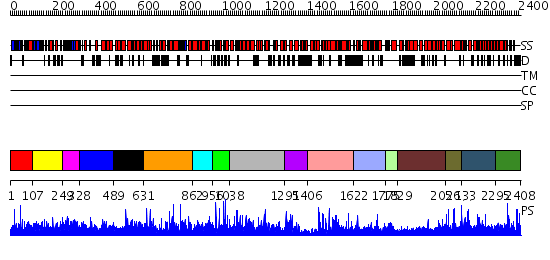
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..248] | 1.013945 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [249..327] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [328..488] | 1.01394 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [489..630] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [631..861] | 1.019952 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [862..955] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [956..1037] | 12.031996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1038..1294] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1295..1405] | 22.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 11 | View Details | [1406..1621] | 19.0 | The crystal structure of Hsp40 Ydj1 | |
| 12 | View Details | [1622..1774] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1775..1828] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1829..2055] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [2056..2132] | N/A | No confident structure predictions are available. | |
| 16 | View Details | [2133..2294] | 1.013962 | View MSA. No confident structure predictions are available. | |
| 17 | View Details | [2295..2408] | 2.014978 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. | |||||||||
| 9 | No functions predicted. | |||||||||
| 10 |
|
|||||||||
| 11 |
|
|||||||||
| 12 | No functions predicted. | |||||||||
| 13 | No functions predicted. | |||||||||
| 14 | No functions predicted. | |||||||||
| 15 | No functions predicted. | |||||||||
| 16 | No functions predicted. | |||||||||
| 17 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)