













| General Information: |
|
| Name(s) found: |
lin-PA /
FBpp0087732
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 858 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
wing disc development
[IMP]
torso signaling pathway [IGI] terminal region determination [IGI] foregut morphogenesis [IMP] epidermis development [IMP] oxidation reduction [IEA] hindgut morphogenesis [IMP] |
| Molecular Function: |
catalytic activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTSSAAGSG AGTGDVCPCS TVATSTTSAS NEQPQTKRQK IEQQIKVGSK ALPPQPPHPP 60
61 DILDLDANSN SHLCSSLSSS SSHSLSTPST ANNSPTTTPC SSRAASYAQL LHNILPQNGD 120
121 TDLHSQPESA HDEVDRAAKL PNLLTLDTTN GNGSHASSRS NCLQATTATS IDDMLEFEQS 180
181 LTRQCLCGVS ERTLRKPFQS HYSQDTNGQK RIAYLREWPT NKLLQFLSNL QLLFDIYLKQ 240
241 NAKGFICTRI MDVCDALIRN DHKLIDEIIV LAGYENSYVQ FLAGRVLAAF LVIAKQELND 300
301 EWLQKIVDQL FNFEQLDQAA VQKIHFSLDI IKRIVEWKDM EIHPLDDDWM ASANSTSAAS 360
361 SVVPLSTEAS VSYMHFPVQE QPLATNYFAL QFREDTEGER ETGQEAPDNR DRHRRHFGED 420
421 MNVAYEPHPA PQSTTMPSGC HVVTLTDSES FDTTHLKCIT IQKLEHKWPT LVKNMSELMA 480
481 PTHQDAAEHC VLNFLQLWEN IISVKANLSI DETRPYYAQL DKFELLLSHS LSCTVYKQML 540
541 CLFNEALCYG STLALQDMLP EETCKLAHQI VCHVRGFRIL ESLPRRQPDN MVSLIGYNGK 600
601 PMVYANGTIT LAHAAQSGDS EEDGAPLDLI EMDKTLLQKM VLLVLKSIAV TVKEIRSDSS 660
661 DSSIDSTDYD AFQDMMLIER SIRDVLSKLE TFIKQTLEFH PECHFSKILI HLFDDQDDHL 720
721 IEAMVCTLDV TSGISFRNNA FPELVAMLNP VYTFLEFLKM TSNSSDLLLD LLVSNETCFL 780
781 LYLLRLLKYI RMNWTMFVHS CHTFGMGSAM LDEAMGVLIR LRLQISRLVS RQLYPYDISP 840
841 VLRLLESCES LYEGNELS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
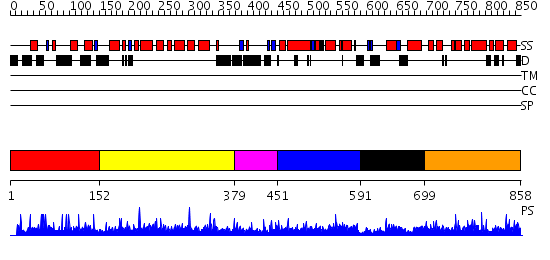
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [152..378] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [379..450] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [451..590] | 1.011982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [591..698] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [699..858] | 3.013974 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.60 |
Source: Reynolds et al. (2008)