













| General Information: |
|
| Name(s) found: |
FBpp0297789
[FlyBase]
FBpp0297785 [FlyBase] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1020 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
inter-male aggressive behavior
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGEVPAETS PVGCNTSNSQ GSSPHPDPIQ QQQLNSSVDS GIAVLEAESP TLRRRQRLHQ 60
61 CQRILQVLQR DQLTHQQLRD RLSKLANKKW RKEESSSEDH ACNVCCADLD LSSAKNYVTC 120
121 CTCEKSVCRG PKCADWRPKD AKWECQLCQS SKESLAHTSS WVAEQMSFNQ HKFVYPMRAR 180
181 SEVYIPIVGD GNDSSMQFES VSQIGQNAHM DERAKIREYV EEIVAEMLGG NLDHVKVGQL 240
241 SKSENYLPAI VNGHTNNSNN NNNNNHADSE LADLSQTRLR SLIETIIAET LRSSTLSASG 300
301 AVSEISLDTR SHVSDLANGN GLKRRHRTEH YFEPKIYQDL LATAVLNKIV DKEGNTRLAA 360
361 ESTPDLSGRH IDENFNAEAL STTSGSSIEP RSDCSLTDHE IGLDNGKSQS LQTDLERESV 420
421 LSDYIAAHMV PLPDFSASVT ESEDDIGSIS SGMIGDGNWE DNWLFKKKRS SATPSSIGML 480
481 VPAPRENVRA QIGDKTTDEV SDLSEIGSDI EESSLDLLRC NDLNDRLLSK HLIGGQNTKM 540
541 VLDELVDRTS LTSHTLPEEN EPAFTETTNE FVVSPMAMPS DIKAPSPTPP PPPMIFQDDL 600
601 LNEEPDHTPI AGSIAEREVK KWYNAVEMPN NPYSPEALKQ RISGTQERYM DVPNISPSAE 660
661 QKALASALTE NPDPPAPKAD YKRYSRDYYI NNAPTSTDST GGGKAATTSA GEDVEDIVIN 720
721 EARKASQTAT EQNPPQESIS VYKATPVQVL DESLDSQSNP SLYSLQTTTT NTSDESDTVR 780
781 IYDFNKQETT VIRAAPAEQQ PSESTTSSME SAQGAPPSVS SSIDSSVSQK RERPVVLQFG 840
841 PGDSVPTIGS PASTPTRGST PPAFRFLQPK RRLIDPSQVL SVDEDDVPEP TTPAAEKPVI 900
901 EDEVAHSMPS VKALAQAFLL TSKHTQPQRR WRAKVRIAAP PDTPDKPNTS LAKRHKLEHA 960
961 VSMAEVADES TIASDLSSLE TDPSIHSEAT PPIASPASPV PVRHGFLRSN IAFFENLKFK1020
1021 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
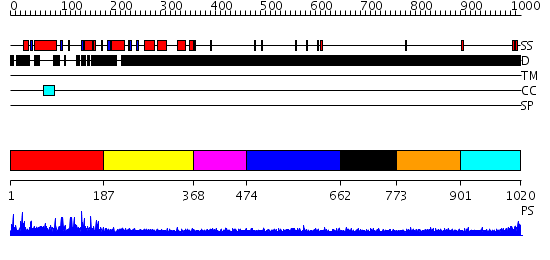
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | 50.69897 | Effector domain of rabphilin-3a | |
| 2 | View Details | [187..367] | 1.122991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [368..473] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [474..661] | 2.152981 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [662..772] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [773..900] | 1.148981 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [901..1020] | 1.124981 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)