













| General Information: |
|
| Name(s) found: |
CG10417-PB /
FBpp0085428
[FlyBase]
CG10417-PA / FBpp0085427 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 662 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein serine/threonine phosphatase complex
[IEA]
|
| Biological Process: |
protein amino acid dephosphorylation
[IEA][NAS]
|
| Molecular Function: |
protein serine/threonine phosphatase activity
[IEA][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGAYLSHPKT DKTSTDQFNE LLAVGASSMQ GWRNSQEDAH NSILNFDNNT SFFAVYDGHG 60
61 GAEVAQYCAD KLPHFLKNLE TYKNGQFEVA LKEAFLGFDK TLLDPSIVSI LKILAGEHNF 120
121 VDAEADDYEE EDLAELQEES NLPLNEVLEK YKGLPQKKDL DLKSSDHKEN FKMRSPYFRG 180
181 RRAAALAAEA TNKAVMDPSA KPDGSSTSAA AAAAALSADG VANSRNPSNV VNPMAGADSN 240
241 TTTSINDLST KNAALKDDSV NDQNEGSNGT DFKHTLVSSS NKKLFATGSN DMTELNQSSK 300
301 NEFTNSSTSK EFERNINSSQ DDEFTDDDAD YEENDNVKSP DTSSAESSDC TENDDDGDED 360
361 GNEDSDEEET DEDQMANDNF CANMIEEPGK DSGCTAVVCL LQGRDLYVAN AGDSRCVISR 420
421 SGQAIEMSID HKPEDDEEAS RIIKAGGRVT LDGRVNGGLN LSRALGDHAY KTNVTLPAEE 480
481 QMISALPDIK KLIITPEDEF MVLACDGIWN YMSSEEVVEF VRCRLKDNKK LSTICEELFD 540
541 NCLAPNTMGD GTGCDNMTAV IVQFKKKLQE LQSTIPPNQT EDKLLKTSEN VSHSLNDQSA 600
601 SKRCASQNAD ADDEILEKNN SKRLKTDLEQ ENIKDRTPSP SNQNEDPTQK AIKEVTIIVS 660
661 SS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
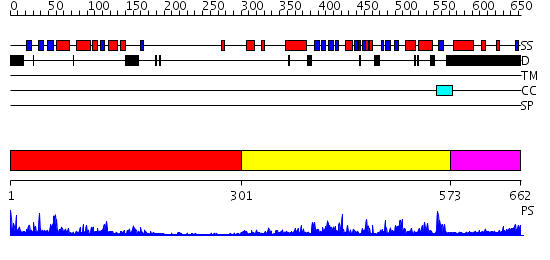
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..300] | 6.61 | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | |
| 2 | View Details | [301..572] | 63.221849 | No description for 2i0oA was found. | |
| 3 | View Details | [573..662] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)