













| General Information: |
|
| Name(s) found: |
p120ctn-PA /
FBpp0110406
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 781 amino acids |
Gene Ontology: |
|
| Cellular Component: |
adherens junction
[IDA][IPI][ISS]
centrosome [IDA] cytoplasm [IDA] |
| Biological Process: |
compound eye morphogenesis
[IGI][IMP]
cell adhesion [IGI] adherens junction maintenance [IMP] |
| Molecular Function: |
binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEARSLKHSL IGLNMNSTHI NMDLSLTHEP ANMQSQFGSF QNYDQFSAAG YSSTPLYITK 60
61 PTGVGAITKV APHCSQNKIE TNSTDNSIPD IENHPLATTV RYVQAGQYED TSEYGVAGLT 120
121 CGIDSEYTAE AVVPYCYTSD ANYTGIQNTT SSIKPFSGRP FNDNINGAEA VADSQCRTFK 180
181 SSAEFKLPQF AMSSINTVPQ RLEEKDDYIE GSENDICSTM RWRDPNLSEV ISFLSNPSSA 240
241 IKANAAAYLQ HLCYMDDPNK QRTRSLGGIP PLVRLLSYDS PEIHKNACGA LRNLSYGRQN 300
301 DENKRGIKNA GGIAALVHLL CRSQETEVKE LVTGVLWNMS SCEDLKRSII DEALVAVVCS 360
361 VIKPHSGWDA VCCGETCFST VFRNASGVLR NVSSAGEHAR GCLRNCEHLV ECLLYVVRTS 420
421 IEKNNIGNKT VENCVCILRN LSYRCQEVDD PNYDKHPFIT PERVIPSSSK GENLGCFGTN 480
481 KKKKEANNSD ALNEYNISAD YSKSSVTYNK LNKGYEQLWQ PEVVQYYLSL LQSCSNPETL 540
541 EAAAGAIQNL SACYWQPSID IRATVRKEKG LPILVELLRM EVDRVVCAVA TALRNLAIDQ 600
601 RNKELIGKYA MRDLVQKLPS GNVQHDQNTS DDTITAVLAT INEVIKKNPE FSRSLLDSGG 660
661 IDRLMNITKR KEKYTSCVLK FASQVLYTMW QHNELRDVYK KNGWKEQDFV SKHFTAHNTP 720
721 PSSPNNVNNT LNRPMASQGR TRYEDRTIQR GTSTLYSAND SSGAVMSNES AMLSEMVRKQ 780
781 L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
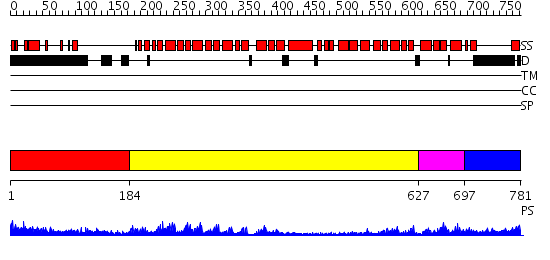
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 42.69897 | Importin alpha | |
| 2 | View Details | [184..626] | 29.69897 | Adaptin beta subunit N-terminal fragment | |
| 3 | View Details | [627..696] | 59.221849 | Structure of the armadillo repeat domain of plakophilin 1 | |
| 4 | View Details | [697..781] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)