













| General Information: |
|
| Name(s) found: |
gi|114581291
[NCBI NR]
gi|114581289 [NCBI NR] gi|114581287 [NCBI NR] gi|114581285 [NCBI NR] gi|114581283 [NCBI NR] gi|114581281 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Pan troglodytes |
| Length: | 727 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFQKAVKGT ILVGGGALAT VLGLSQFAHY RRKQMNLAYV KAADCISEPV NREPPSREAQ 60
61 LLTLQNTSEF DILVIGGGAT GSGCALDAVT RGLKTALVER DDFSSGTSSR STKLIHGGVR 120
121 YLQKAIMKLD IEQYRMVKEA LHERANLLEI APHLSAPLPI MLPVYKWWQL PYYWVGIKLY 180
181 DLVAGSNCLK SSYVLSKSRA LEHFPMLQKD KLVGAIVYYD GQHNDARMNL AIALTAARYG 240
241 AATANYMEVV SLLKKTDPQT GKVRVSGARC KDVLTGQEFD VRAKCVINAT GPFTDSVRKM 300
301 DDKDAAAICQ PSAGVHIVMP GYYSPESMGL LDPATSDGRV IFFLPWQKMT IAGTTDTPTD 360
361 VTHHPIPSEE DINFILNEVR NYLSCDVEVR RGDVLAAWSG IRPLVTDPKS ADTQSISRNH 420
421 VVDISESGLI TIAGGKWTTY RSMAEDTINA AVKTHNLKAG PSRTVGLFLQ GGKDWSPTLY 480
481 IRLVQDYGLE SEVAQHLAAT YGDKAFEVAK MASVTGKRWP IVGVRLVSEF PYIEAEVKYG 540
541 IKEYACTAVD MISRRTRLAF LNVQAAEEAL PRIVELMGRE LNWDDYKKQE QLETARKFLY 600
601 YEMGYKSRSE QLTDRSEISL LPSDIDRYKK RFHKFDADQK GFITIVDVQR VLESINVQMD 660
661 ENTLHEILNE VDLNKNGQVE LNEFLQLMSA IQKGRVSGSR LAILMKTAEE NLDRRVPIPV 720
721 DRSCGGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
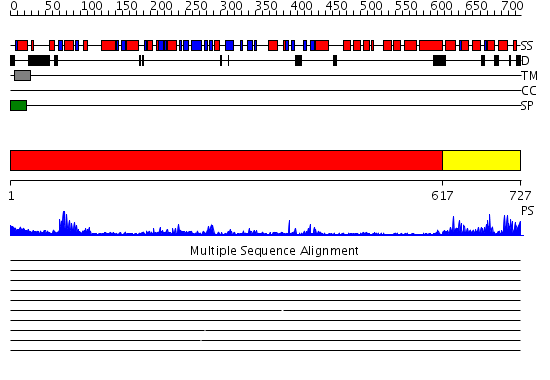
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..616] | 21.0 | L-aspartate oxidase | |
| 2 | View Details | [617..727] | 39.69897 | CALMODULIN, NMR, 30 STRUCTURES |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|