













| General Information: |
|
| Name(s) found: |
gi|125561987
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Oryza sativa Indica Group |
| Length: | 633 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVAVPDHTF NNGYSSKNQI EGRSLSWKRV FVQTDKGSVL GIELERGENA HTVKKKLQLA 60
61 LNIPTEESSL TCGDQLLNND LSYICNDSPL LLTRNHMHRS CSTPCLSPNG KDVQHCDDSR 120
121 VIEIVGCTSP SARMKQLVDD IVRGIEKGIE PVAISSGMGG AYYFRDMWGE HAAIVKPTDE 180
181 EPFGPNNPKG FVGKSLGLPG LKKSVRVGET GSREVAAYLL DHKNFANVPP TMLVKITHSV 240
241 FHMNEGVDYK TKSSDNKTQA FSKLASLQEF IPHDYDASDH GTSSFPVSAV HRIGILDIRI 300
301 FNTDRHAGNI LVRKLYNDAS RFETQTELIP IDHGLCLPES LEDPYFEWIH WPQASIPFSE 360
361 EDLEYITNLD PIKDAEMLRM ELHTIHEASL RVLVLSTTFL KEAAACGFCL SEIGEMMSRQ 420
421 FTRKEEEPSD LEVLCMEARN WVEEREWLLP QADFEGEDDN ESTQFDLDSE DDSTTFEASF 480
481 SNNIRPIKGN SRDPPSKLAE VNEYVDEDDN NEFNKDDVGT CTSPITTWTP STSNLSISSN 540
541 ELSFSGRRKS HSGVAKNKVT SKINSNSYSG NHSAKEKLPH NSSFAKLSDL SANKWSPFLE 600
601 KFQDLLQSMF QDRKQTAGRN PWLTQRLGTS CQF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
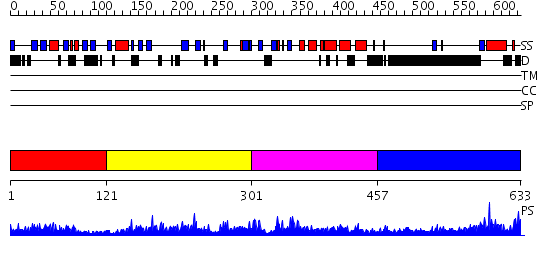
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 24.69897 | Solution Structure of S5a UIM-1/Ubiquitin Complex | |
| 2 | View Details | [121..300] | 1.084969 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [301..456] | 4.081966 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [457..633] | 1.037958 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)