













| General Information: |
|
| Name(s) found: |
gi|168278070
[NCBI NR]
gi|123999981 [NCBI NR] gi|123993665 [NCBI NR] |
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 636 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNPSAPSYPM ASLYVGDLHP DVTEAMLYEK FSPAGPILSI RVCRDMITRR SLGYAYVNFQ 60
61 QPADAERALD TMNFDVIKGK PVRIMWSQRD PSLRKSGVGN IFIKNLDKSI DNKALYDTFS 120
121 AFGNILSCKV VCDENGSKGY GFVHFETQEA AERAIEKMNG MLLNDRKVFV GRFKSRKERE 180
181 AELGARAKEF TNVYIKNFGE DMDDERLKDL FGKFGPALSV KVMTDESGKS KGFGFVSFER 240
241 HEDAQKAVDE MNGKELNGKQ IYVGRAQKKV ERQTELKRKF EQMKQDRITR YQGVNLYVKN 300
301 LDDGIDDERL RKEFSPFGTI TSAKVMMEGG RSKGFGFVCF SSPEEATKAV TEMNGRIVAT 360
361 KPLYVALAQR KEERQAHLTN QYMQRMASVR AVPNPVINPY QPAPPSGYFM AAIPQTQNRA 420
421 AYYPPSQIAQ LRPSPRWTAQ GARPHPFQNM PGAIRPAAPR PPFSTMRPAS SQVPRVMSTQ 480
481 RVANTSTQTM GPRPAAAAAA ATPAVRTVPQ YKYAAGVRNP QQHLNAQPQV TMQQPAVHVQ 540
541 GQEPLTASML ASAPPQEQKQ MLGERLFPLI QAMHPTLAGK ITGMLLEIDN SELLHMLESP 600
601 ESLRSKVDEA VAVLQAHQAK EAAQKAVNSA TGVPTV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
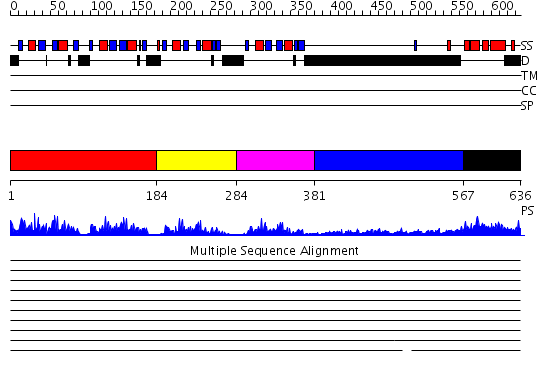
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 59.69897 | Poly(A)-binding protein | |
| 2 | View Details | [184..283] | 5.3 | No description for 2do0A was found. | |
| 3 | View Details | [284..380] | 19.522879 | No description for 2d9pA was found. | |
| 4 | View Details | [381..566] | 4.69897 | Sec24 | |
| 5 | View Details | [567..636] | 44.522879 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)