













| General Information: |
|
| Name(s) found: |
gi|114645574
[NCBI NR]
gi|114645572 [NCBI NR] gi|114645570 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Pan troglodytes |
| Length: | 706 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKIFSESHKT VFVVDHCPYM AESCRQHVEF DMLVKNRTQG IIPLAPISKS LWTCSVESSM 60
61 EYCRIMYDIF PFKKLVNFIV SDSGAHVLNS WTQEDQNLQE LMAALAAVGP PNPRADPECC 120
121 SILHGLVAAV ETLCKITEYQ HEARTLLMEN AERVGNRGRI ICITNAKSDS HVRMLEDCVQ 180
181 ETIHEHNKLA ANSDHLMQIQ KCELVLIHTY PVGEDSLVSD RSKKELSPVL TSEVHSVRAG 240
241 RHLATKLNIL VQQHFDLAST TITNIPMKEE QHANTSANYD VELLHHKDAH VDFLKSGDSH 300
301 LGGGSREGSF KETITLKWCT PRTNNIELHY CTGAYRISPV DVNSRPSSCL TNFLLNGRSV 360
361 LLEQPRKSGS KVISHMLSSH GGEIFLHVLS SSRSILEDPP SISEGCGGRV TDYRITDFGE 420
421 FMRENRLTPF LDPRYKIDGS LEVPLERAKD QLEKHTRYWP MIISQTTIFN MQAVVPLASV 480
481 IVKESLTEED VLNCQKTIYN LVDMERKNDP LPISTVGTRG KGPKRDEQYR IMWNELETLV 540
541 RAHINNSEKH QRVLECLMAC RSKPPEEEER KKRGRKREDK EDKSEKAVKD YEQEKSWQDS 600
601 ERLKGILERG KEELAEAEII KDSPDSPEPP NKKPLVEMDE TPQVEKSKGP VSLLSLWSNR 660
661 INTANSRKHQ EFAGRLNSVN NRAELYQHLK EENGMETTEN GKASRQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
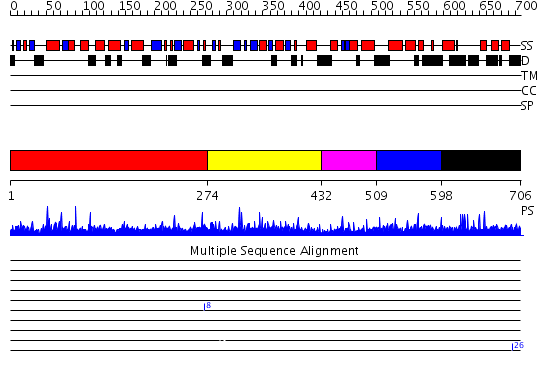
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..273] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [274..431] | 1.015925 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [432..508] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [509..597] | 1.017978 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [598..706] | 1.011995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)