













| General Information: |
|
| Name(s) found: |
gi|114684644
[NCBI NR]
gi|114684642 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Pan troglodytes |
| Length: | 780 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVDLEKLR ASGAGKAIGV LTSGGDAQGM NAAVRAVTRM GIYVGAKVFL IYEGYEGLVE 60
61 GGENIKQANW LSVSNIIQLG GTIIGSARCK AFTTREGRRA AAYNLVQHGI TNLCVIGGDG 120
121 SLTGANIFRS EWGSLLEELV AEGKISETTA RTYSHLNIAG LVGSIDNDFC GTDMTIGTDS 180
181 ALHRIMEVID AITTTAQSHQ RTFVLEVMGR HCGYLALVSA LASGADWLFI PEAPPEDGWE 240
241 NFMCERLGET RSRGSRLNII IIAEGAIDRN GKPISSSYVK DLVVQRLGFD TRVTVLGHVQ 300
301 RGGTPSAFDR ILSSKMGMEA VMALLEATPD TPACVVTLSG NQSVRLPLME CVQMTKEVQK 360
361 AMDDKRFDEA TQLRGGSFEN NWNIYKLLAH QKPPKEKSNF SLAILNVGAP AAGMNAAVRS 420
421 AVRTGISHGH TVYVVHDGFE GLAKGQVQEV GWHDVAGWLG RGGSMLGTKR TLPKGQLESI 480
481 VENIRIYGIH ALLVVGGFEA YEGVLQLVEA RGRYEELCIV MCVIPATISN NVPGTDFSLG 540
541 SDTAVNAAME SCDRIKQSAS GTKRRVFIVE TMGGYCGYLA TVTGIAVGAD AAYVFEDPFN 600
601 IHDLKVNVEH MTEKMKTDIQ RGLVLRNEKC HDYYTTEFLY NLYSSEGKGV FDCRTNVLGH 660
661 LQQGGAPTPF DRNYGTKLGV KAMLWLSEKL REVYRKGRVF ANAPDSACVI GLKKKAVAFS 720
721 PVTELKKDTD FEHRMPREQW WLSLRLMLKM LAQYRISMAA YVSGELEHVT RRTLSMDKGF 780
781 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
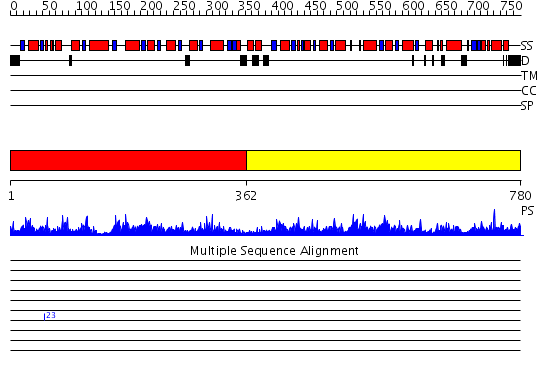
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..361] | 106.0 | ATP-dependent phosphofructokinase | |
| 2 | View Details | [362..780] | 77.0 | No description for 2higA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)