













| General Information: |
|
| Name(s) found: |
UTP22_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1097 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNGLKREHES SSSQDGSKTP ETEYDSHVDS IEDIHSLASK RKKLNEKKEN LEDLTLLKTS 60
61 AFELKLNELI REISVRGKYF RHANTFVEKI KDLIFKTPVI PETNFWSACK NLEKDKKVIV 120
121 PLAEPLSAKD TNLRASFVPP KTVTPGIFSC SNKFFLNPDG WSYDLFLEIP ESIFTQKDYL 180
181 NGRYFRKRAF YLTCIAKHLL ENLGNEVKLE FVAFNDDIRR PILAILPESK GFAATGKRFT 240
241 VFLIPTVRQI FPVSKLLPHK NAIRDFMEHE ELKPTPFYNN SVLEEQNLLF YRDLVKKYSV 300
301 NPQFLDACGL GSTWLNMRGF SSSIHSNGFG LLEWYVLMAL LMSSTGLPAG NVLNTYLTAA 360
361 QFFKSMLQFL SSKNLTSTLF KLNADSSNLK IGNGHLPTLI DCNTGFNLLG KMKQSFFEYF 420
421 QASCRHTLNL LDENANYNFS KIFITHVNVP ALEFDVSGCI PLEPKELEDP NFCRKTDLDS 480
481 PYSLYLEYTW DLLQHALGDR VCQIILYSSI CTSCSINESL KTKLPKLISF GLLLNPDALL 540
541 RLVDIGPSPD DTVGSQKFRE FWGEVSELRK FKNGSIAESV YWECSSPDER IRIPQRIIRH 600
601 ILNRHLGNNV GDRVSFRNEK FRVYVHSKIS PNTDTYNEYV PVMEAYNEAV KSLINLSDIP 660
661 LSIAEILPAD ESLRYSSSSV PFYESSTCAP IDVVFQFESS SKWPDELEGI QRTKIAFLLK 720
721 IAELLEALDN VERASVGLEN TDNPTHNCCF LQVLFSNNFT FRYRLRNDRE IFFWKSLERN 780
781 PSTKLSAQKG LYAYEHMFQF IPRHTLAIQA ICQAHRSYSM AVRLAKHWFY SHLLTDHVTD 840
841 EVIELLVASV YINSSSWRTT SSGETSFCRM LHFLAHWDWR FDPLIINSNG KLPHDVRHQA 900
901 TEKLESIRKQ DVAIAHNAYY IITDYDFDGN HIGYYKPSKI IANRITSLAR ASLSELLKDT 960
961 PNYKSIFKSS LDIYHVVIDV NINKLPMYRE SNLTKYKNLQ NMSSERPGFE PITEFVKELH1020
1021 RCFEDTISFF YNKKNPKVVT GVFNPRILAN RPYRTNIDYP FKVVDKDTVV LDADVVCEEI1080
1081 RQVGGDLIRS IQLQLKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
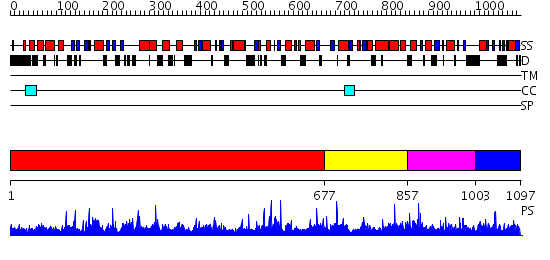
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..676] | 2.55 | Crystal structure of poly(A) polymerase in complex with 3'-dATP and magnesium chloride | |
| 2 | View Details | [677..856] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [857..1002] | 2.049975 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1003..1097] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)