













| General Information: |
|
| Name(s) found: |
KLF1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 781 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKAKKSRAY QEGDIRYKCD FQGCTKSFTR KEHARRHFRS HTNSKAFICP HCSSSFTRSD 60
61 VLNRHVNQKH VKQNDAGSQT HPKHDKTLIS SSDQELPFPS YVEADVQDAA YTDLKSHIDQ 120
121 PILPDQPIIS DNFNEGLQSA IAQTGNNYMF TTSRRNNSIS GSITIPESDQ QINLQNKFLS 180
181 TPSIKNNANK SLSPQDSISI FGTSPFNAYQ QNTDNFVCWL FDNMEKDAPT ESVSNFTDGI 240
241 NAKQLNLMPV LDSPTIDFLS SHFRVDDKLV AKDLLSLSAY SSLIQVMNNN YQLSEEALQY 300
301 ITRSRVNLWI AHYWKDFHPR WPFLHRGTLK VDEAPVELLL AMITMGMHFV GDAFAYSIAV 360
361 SIHSTLRFSI YTHPDFKPPA SLWVYQALLI AEIFEKMTST VEQHNLSQIF HGVTIESMQN 420
421 GLSSKDTVTE KIPKNMTGTN IAQHKWHQWV DREASKRIAF FSFVLDSQHV ILFGYRPLID 480
481 ITSVGLPLLC DEALWNADSY EAWTSLLNEN DPPHFFPVLK MFLTNEHLPP KLSPWNMMIV 540
541 LHGLTTIGWI LSRENLGIVE NIMQNNGTNL KNWRILLKAS YKFWLRTYRV FFLNNGTLPL 600
601 NHPYVRGCLA TYELAHISLH TNIVALQTYA KSIVSTSRRL YASTSRYVSA WLASDDSEVS 660
661 IKHAVDMVEA FLGGDMEYDV QNETGLHRPW CLYVSTLILW AYGYVSDGRC EMLEPGNNDC 720
721 KSQLNAYLMQ MRTGLSEKAK DHVYVRSKTL PLLKCVIDVL CPARWGLLVD GVRILSKLVQ 780
781 L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
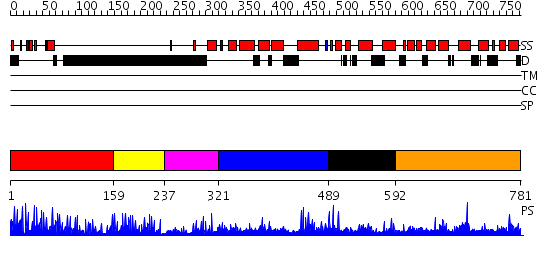
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 52.69897 | No description for 2i13A was found. | |
| 2 | View Details | [159..236] | 6.488993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [237..320] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [321..488] | 1.164993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [489..591] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [592..781] | 1.028993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)