













| General Information: |
|
| Name(s) found: |
YKR3_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 797 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNEYVDMNS SSEDSDGDSI LEEGRLRPSF RGQQKERDML GIFGEEDEDG FHNSGIGSAR 60
61 LRRKNISFVE KSEQVKANKQ VTADDLLEAH SIPQLKNKND EVSQAKNIPK MKFNTTGFGA 120
121 KMLEKMGYKQ GQGLGANAEG IAEPVQSKLR PERVGLGAVR ERTEKQRKEA IARGEISDSE 180
181 DEKHTVKQKP LREKKKKPLK SSEEISKDMG SYNLPRFLAS LIDASLNDTK EIEFVTSNKE 240
241 ELGLEGRDMS TSGINQLSRL ARVECEHHAS AWQQLQARRA YVKMELKRVT TEFDEKSVEI 300
301 SRLEKLLGKV MEVKSRSMEF TVPEAEIDVI EKRLQPLNNL IETLPVEFSE ASMHFELDSV 360
361 AVSILAFVLS EPIKNWDVWK HPYFMLESFL SWKNSLYSKD FRPKREESST FMDIDVEFDD 420
421 ELEGQSLTHY ESFMMFVWKK KIGEELKKWI IQDSLKALQL LEAWDPVVPE KVKDSLIQDD 480
481 ILPRLKDAVS KWNPKLKLKK NDSLHHCIFP WLPYLEKHAD SLLQSVLVQF SLILSPWKIK 540
541 NGSIDDFSVW RSAFANDALD RLLEKVILPK LEKLMDEELV IDPSNQDLEI FFIILSWKGS 600
601 FKAMVFGQLF ADHFFPKWLE TLYQWLTEAP NFDEASEWYT WWKSVFPKDL LSNAYIQQGF 660
661 SKGLDMMNEC LENKSITAPL PFAKDSTKGV NLQFSKEKHE FTAESDDTTS YDEPLVSFRE 720
721 LVEEFCAENS LLFVPLRRSH LSTGSALFRI STQASKARGI TVYLRNDIIW KKSPGASEDT 780
781 PYDPIGFNEI LLMFNNN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
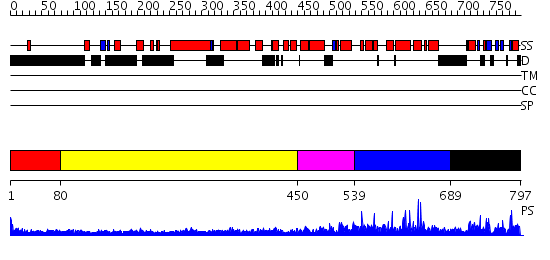
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [80..449] | 20.69897 | Heavy meromyosin subfragment | |
| 3 | View Details | [450..538] | 6.522879 | No description for 2i1jA was found. | |
| 4 | View Details | [539..688] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [689..797] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)