













| General Information: |
|
| Name(s) found: |
NU132_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1162 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSILGKRKN EEVVSFSPLK RISVEKSLLD STAYNNSLDW LRSKNFKVSC LLKHFNSKII 60
61 NDHPLSGSCY TDIGYALINS RKACFILSYR QSLGTAEPPT ITFPLPEEDS NGFSGQNALT 120
121 AFVPSDASDK EPGLLIVMPI SGRIAYWTSI GNALAQSYIC PQGMESLIKL LPKEKCEHLC 180
181 CSNPMKFIIS TNFGRLFSVQ LRDPAGQPDV SVQLFASDIS TFSTILQKMK IFNYPSIHII 240
241 ALKSPPLFSP YQHLLYVAEA SGLLEIYDLK LENKLVSGMN LSPIFKQVLR EGCPDASGLE 300
301 VLDLTICPTN GNLVSFLVCW KNSINYRYMI ISLDFSDISS PSVMNIHPLY SFSSKSLESS 360
361 KLHYSSSGNS LFVVLTDAVI IVHVQEDDKD IVSRTSWEEV IRMNTNVSGG IFMSTCYKYV 420
421 LGKYSIPTES CFIATPYSGI AEIEVHSLEH PANNESLVKS KLEEAVFYSF LPGNPIDFSC 480
481 NYLRSIKKPE LERIIVDLGM DILNSRSTHL PPLFASLMQH LSCRLNSLNN LVRYIRSMSL 540
541 DVDRQVLYKL RVMGEKCNSV RYLWNTIDTE FSTVSHSLIF QRIIYRLTQS ASSDNALREW 600
601 FLHNIESIDQ LIAQAHEFCI DSGSRVQELP LEVLDVIMEA NEVILAIQSS ALAYRRESQK 660
661 IYKLSIDTFG EEVPWTSTPE TLVLLCRQFE LTRSALVQSH QGTSDVENTF KIKDKGVLRN 720
721 VVSNLEVQLV ALTEVCFDAY SERIRWIEQR CGKDASEIQD VKEAFAVNRR FWVQTLSDIG 780
781 KGSSAIRIAE KYSDYRSLVE LCYQLYEDNE LTDALNNYLD LFGIKFAFIL YDYFVENGMA 840
841 LELLNSDRFN KSYLKQFFKS RDYNQISWMH DMRLGDYDAA SHRLLQLATK QEKLVDKKES 900
901 ELSLSKLFLY AVPSNSGNIR DLVLVEQKLE QLHIQKMVSK SVMPVVERLR SQGKKYQLVE 960
961 AVVDDLIGAK VAPVIARQVM QRVVKKFIAG QVVEATELLE YLSFSLYRRE DLVEGEVTDY1020
1021 YLALRLLLTT RLTDDAKRFY ENTIWRRAVL HDNWIQVLDT QGKNDAIIET QFRMSALYRT1080
1081 LEAVTINGLF HEGLIRPGSL SSCKFEGYDP QNLISIYPPA RFGDVTEVTK VLNRESVKLD1140
1141 HYLTKTNLNT CYISMCLSCD TI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
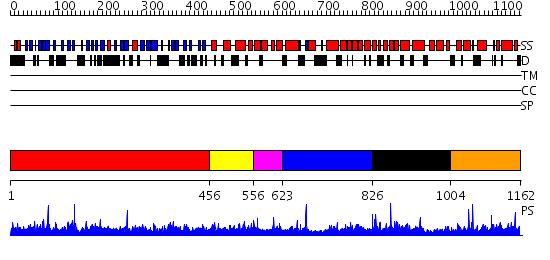
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..455] | 97.0 | The crystal structure of the N-terminal domain of Nup133 reveals a beta-propeller fold common to several nucleoporins | |
| 2 | View Details | [456..555] | 1.050992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [556..622] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [623..825] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [826..1003] | 3.074977 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1004..1162] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)