













| General Information: |
|
| Name(s) found: |
ATG13_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRLNTQLPR MYSAPPGHSK AVSTELNKDL SSVGGRSAKL GQVIHHCFYK TGLIILESRL 60
61 NVFGTSRPRE SSKNNKWFNL EIVETELYAE QFKIWKNIEL SPSRKIPPMV LHTYLDISDL 120
121 SKNQTLSVSD GTHSHAINFN NMSTMKIVLE RWIVNLDGEA LSTPLELAVL YKKLVVLFRS 180
181 LYTYTHLMPL WKLKSKIHKL RAHGTSLKVG CALSTDDVLS NDFLPISAPI SSSLGSSIAT 240
241 FSFSPVGTPA GDFRISVQYR KNCHFEVHDS DALLSNQLLS ADKHQLAASN NSQDFEDGKQ 300
301 YDQPPPSFAT RLAKQSDPNS LLQSEIQHLA SIESITAQAA PLVTIHPFKS PSLSASPGSN 360
361 FDNMSISPKV AVNRYIHRGP SATSLNKFSM ISDAASKSRA KLPPLTSGSL KLNTLDISNT 420
421 PNLRRFSSSF GPRERKESFS SRNRLPLVNH PIRSIFKHNV SENPITDHSE HAVYDSEFAS 480
481 KDDLSGFIQL LDSHAHHLNA SEGSKSSGSF PGKVQTLTSG ISPVAHPHNS LGSSNEIFDI 540
541 DTYNHSIDNS GSRFTEAVKH NLGNSSHSIM RHHTLGTLRS RPSFSEKSTF PAPLTSISQA 600
601 STFQGDNRSP STVIPHTQTE VPSANDTSKQ LASLHDMRKS QSPICARSAT SAGLPRFEYH 660
661 TSLSKSLEHS STPASLQATK TPSPSFVLEP GIPQEYKKHF DNLSEERRQC LTPSTPTYEY 720
721 YNEHNPNYDD DLLFTMTDMT LEPHDVSAIR LGSPKSDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
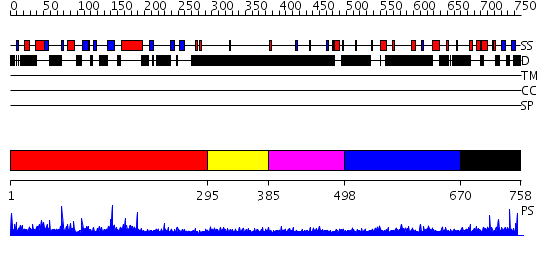
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..294] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [295..384] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [385..497] | 3.17599 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [498..669] | 3.177994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [670..758] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)