













| General Information: |
|
| Name(s) found: |
YDM6_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 929 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARKGKVNTL PQPQGKHQRK GKKQLENKIL HSYEEESAGF DSEELEDNDE QGYSFGVNSE 60
61 DDEEIDSDEA FDEEDEKRFA DWSFNASKSG KSNKDHKNLN NTKEISLNEE DDSDDSVNSD 120
121 KLENEGSVGS SIDENELVDL DTLLDNDQPE KNESNTASTI RPPWIGNNDH ATDKENLLES 180
181 DASSSNDSES ELTDSADNMN ESDSESEIES SDSDHDDGEN SDSKLDNLRN YIVSLNQKRK 240
241 KDEADAESVL SSDDNDSIEE ISIKKVKYDP HETNKESEYN LIGSSEKTID ITDLLDSIPM 300
301 NEQLKVSLKP LVSESSSISS KKLDAPLAKS IQDRLERQAA YEQTKNDLEK WKPIVADNRK 360
361 SDQLIFPMNE TARPVPSNNG LASSFEPRTE SERKMHQALL DAGLENESAL KKQEELALNK 420
421 LSVEEVAERT RQLRFMRELM FREERKAKRV AKIKSKTYRK IRKNRKEKEM ALIPKSEEDL 480
481 ENERIKSEEA RALERMTQRH KNTSSWTRKM LERASHGEGT REAVNEQIRK GDELMQRIHG 540
541 KEISEMDGED VSEFSDSDYD TNEQVSTAFE KIRNEEEPKL KGVLGMKFMR DASNRQKALV 600
601 QDEMQAFEDE LAGVPNEDDT SQKGEDGVPG VLIGNNTGRR SFKPSEEAAK LSLPSRKNPF 660
661 VSDSAVLKVN KPEMKEGQKK AEARKKKESP LEATEETNPW LQVPDQRTSS AKKLDKNSSK 720
721 ADKKNHKLKM DKVASLQELV EEPKVQPDLI FEEKAFESAS EAESDVDVSV PMLKPTKGRL 780
781 SIKQRELVAK AFAGDDVVAE FEKDKEDWVQ EDAPKEEDHS LPGWGSWGGV GVKQRKTKPK 840
841 VKKIAGLDPS KRKDSKLKHV IINEKRNKKA AKLTADSVPF PFESREQYER SLNLPMGPEW 900
901 TTRASHHKAV APRVVTKRGK VINPIKAPN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
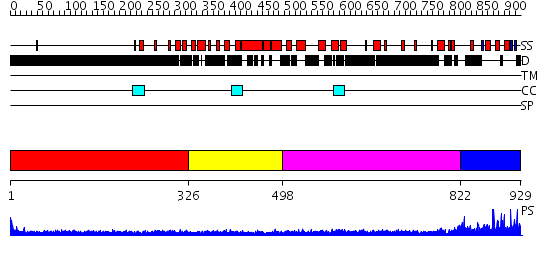
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 15.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [326..497] | 1.17 | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | |
| 3 | View Details | [498..821] | 7.522879 | Tropomyosin | |
| 4 | View Details | [822..929] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)