













| General Information: |
|
| Name(s) found: |
PLD1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 1369 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIHQGGNSV IDNVPYSLNT NVNDSIYSEK GTGRKDAEDH TPSKITDLEK NVDHSIPSFP 60
61 ENDPKNYSEF VNLNPPKRPD LEHTRGSSWH TASENVNDLA ANDSTRVQTP EFITQTMEDE 120
121 NVEVPPLERD ERDAAAAHTS KANRNSARQM WAQLMASVRK FKREDEKPIL KENLPAINLF 180
181 EAGIPASLPI AKHFIRDKSG QPVLPIITDL IKVSVLDVEP KHNRIHSTFT IQVEYGTGPH 240
241 AIRWLIYRQL RDFINLHSHF LFFEFQHRFS GRRMKLPKFP KEVLPYLVKL RGYQKILYSN 300
301 PSDQLIDETH SISDISWESH SQDGDRTTGQ PRHANNGRKK HGNFWTIQGN TLGVYLQEMI 360
361 HNLQFFPEVN VLYSFLEFSS LGLYLAGAGT FHGKEGFATL KRNYSPTQYM LCCNTTMMKT 420
421 RSQPFWIIVS ESCIILCDNM LSMQPADVFI WDVDFEITRK NFRKAHSKDT NEKIRLSHHS 480
481 FKIKNRQKVM KLSVRSGRWL QQFINSVQVA QGLTAWCEIH RFDSFAPVRT NVAVQWMVDA 540
541 RDHMWNVSRA IKNAKRCIMI HGWWLSPELQ MRRPYSMAHK WRIDRILNEK AHEGVMVYIM 600
601 IYRNIDATIP IDSFHTKEHL QSLHPNIYVI RSPSHFRQNA LFWAHHEKLV VVDDAITFIG 660
661 GIDLCFGRYD TPQHILYDDK PVADKTGLCE QTWRGKDYSN ARVHDFFDLT EPYKDMYDRL 720
721 AVPRMGWHDV SMCIIGQPAR DAARHFVQRW NYLIQCKKPA RKTPLLIPPP DFTTDQLTDS 780
781 QLTGTCEVQV LRSAGLWSLG LVDTVEQSIQ NAYVTCIEKS EHFIYIENQF FVTSTTCEGT 840
841 TIENRVGDAL VERIIRAHKN NEKWRGVIMI PLLPGFEGQI DLQEGGSLRL IVECQYRSIC 900
901 HGEHSIFGRL NAKGIDGSKY LRFYGLRGWA HLGENHELVT EMIYVHAKIL IADDRVAVIG 960
961 SANINERSLL GNRDSEIAAV IRDTLTIDSK MDGKPYKVGK FAHTLRKRLM REHLGLETDV1020
1021 LEQREYNMDG LDRDTEWKRV EVWTPDEGNA INGSAYTAEE LKMKYRSQSQ FTTTPDILRK1080
1081 AEKSMKKLDQ RVSLIPSSIE FNIKTQKDKV EFEKNYEKSK KGPDVIANAL VGGIPLSLKT1140
1141 KEDSLYELSK FSQCGEDQRP MVLKDPDHLV PEPSRPHCGN GLVFYDDIPL LEVNPISGET1200
1201 IPKFDASSFE DPVCDEFFED IWSKVASNNT TIYRHIFRCV PDDEMLTWES YNEWKKYGKR1260
1261 FKEEQARWRQ EELSNLHETH EKSENDPKNP KAGSQGSGNT SASEDSKTEK PKTRTNNGLQ1320
1321 VPDKRVVYDL LRGIRGQLVE LPLKWMSTES NARNWLSSID KIPPLEIYD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
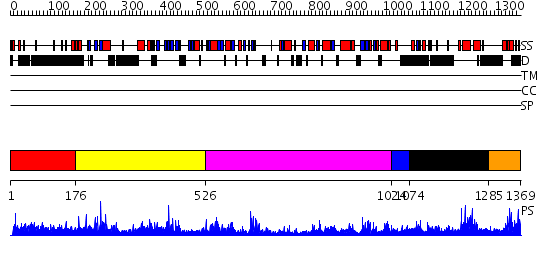
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] | 4.037995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [176..525] | 3.44 | Crystal Structure of a N-terminal Fragment of SKAP-Hom Containing Both the Helical Dimerization Domain and the PH Domain | |
| 3 | View Details | [526..1023] | 36.154902 | Phospholipase D | |
| 4 | View Details | [1024..1073] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1074..1284] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1285..1369] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)