













| General Information: |
|
| Name(s) found: |
PFKA_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 942 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGETVHGIS CYSVVANTED TYNQTLDFYQ KLGFKKVASF GTSDSDNARV CNESLREDWM 60
61 HVAGNNSAES VTIKFRLVPG ELSLSPAAED SEWRGQKSSL VFYYPNLLDL LKQLSADAIK 120
121 YQAFPNEKKP DEVYVEDPLG NLIGFSDRYN PFAHANLKKS EESGAASNLE SGLATPVVET 180
181 LKKATTSDKP AGVKKKIAVM TSGGDSPGMN AVVRAVARIA IHRGCDAFAI YEGYEGLVQG 240
241 GDMIKQLQWG DVRGWLAEGG TLIGTARCMA FRERPGRLRA AKNLISAGID SIIVCGGDGS 300
301 LTGADIFRSD WPGLVKELED TKAITPEQAK LYRHLTIVGL VGSIDNDMSS TDVTIGAFSS 360
361 LHRICEAVDS ISSTAISHSR AFIVEVMGRH CGWLAVLAAL ATGADFVFIP ERPAEVGKWQ 420
421 DELCNSLSSV RKLGKRKSIV IVAEGAIDSE LNHISPEDIK NLLVERLHLD TRVTTLGHVQ 480
481 RGGIPCAYDR MLATLQGVDA VDAVLASTPD TPSPMIAING NKINRKPLME AVKLTHEVAD 540
541 AIEKKQFAHA MELRDPEFAD YLHTWEGTTF IEDESHFVPK DERMRVAIIH VGAPAGGMNS 600
601 ATRAAVRYCL NRGHTPLAID NGFSGFLRHD SIHELSWIDV DEWCIRGGSE IGTNRDTPDL 660
661 DMGFTAFKFQ QHKIDALIII GGFEAFTALS QLESARVNYP SFRIPMAIIP ATISNNVPGT 720
721 EFSLGCDTCL NAVMEYCDTI KQSASASRRR VFVCEVQGGR SGYIATVGGL ITGASAIYTP 780
781 EDGISLDMLR KDIDHLKATF ALEAGRNRAG QLILRNECAS KVYTTEVIGN IISEEAHKRF 840
841 SARTAVPGHV QQGGNPTPMD RARAARLAMR AIRFFETCRA NDLGNDPSSA VVIGIRGTGV 900
901 SFSSVADVEN NETEIEMRRP KNAWWRDMHN LVNILAGKTF AD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
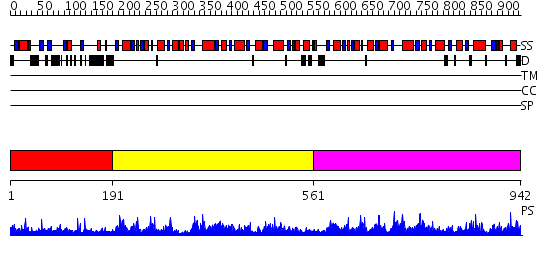
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 2.5 | Crystal Structure of the Glyoxalase Family Protein APC24694 from Bacillus cereus | |
| 2 | View Details | [191..560] | 111.0 | PHOSPHOFRUCTOKINASE. STRUCTURE AND CONTROL | |
| 3 | View Details | [561..942] | 76.39794 | No description for 2higA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)