













| General Information: |
|
| Name(s) found: |
SULH2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 840 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRAWGWVRNK FSSEDDYNDG ASNKDYPDRF NEFDNSQNDH NDYTQNNAQF QNAQTTTFGR 60
61 TISRVKAYYE IPEDDELDEL ASIPQWFKKN VTSNIFKNFL HYLKSLFPII EWLPNYNPYW 120
121 LINDLIAGIT VGCVVVPQGM SYAKVATLPS EYGLYSSFVG VAIYCFFATS KDVSIGPVAV 180
181 MSLITAKVIA NVMAKDETYT APQIATCLAL LAGAITCGIG LLRLGFIIEF IPVPAVAGFT 240
241 TGSALNILSG QVPALMGYKN KVTAKATYMV IIQSLKHLPD TTVDAAFGLV SLFILFFTKY 300
301 MCQYLGKRYP RWQQAFFLTN TLRSAVVVIV GTAISYAICK HHRSDPPISI IKTVPRGFQH 360
361 VGVPLITKKL CRDLASELPV SVIVLLLEHI SIAKSFGRVN DYRIVPDQEL IAMGVTNLIG 420
421 IFFNAYPATG SFSRSAIKAK AGVKTPIAGI FTAAVVILSL YCLTDAFYYI PNAILSAVII 480
481 HAVTDLILPM KQTILFWRLQ PLEACIFFIS VIVSVFSSIE NGIYVSVCLA AALLLLRIAK 540
541 PHGSFLGKIQ AANKYGSDNI ANVRDIYVPL EMKEENPNLE IQSPPPGVFI FRLQESFTYP 600
601 NASRVSTMIS RRIKDLTRRG IDNIYVKDID RPWNVPRQRK KKENSEIEDL RPLLQAIIFD 660
661 FSAVNNLDTT AVQSLIDIRK ELEIYANETV EFHFTNIRSG WIKRTLVAAG FGKPKGHAVD 720
721 ASVCVEVAAP LRDANLSAES SRNLSRIITP IYDDEEGNVS GHIYELDGKN NSDLSMHCQK 780
781 GSNQVEIEFV EFNSRKYPFF HVDVASAVVD LQHRLLSPQK SDSFGSLKEG GTTTIKKIEN 840
841 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
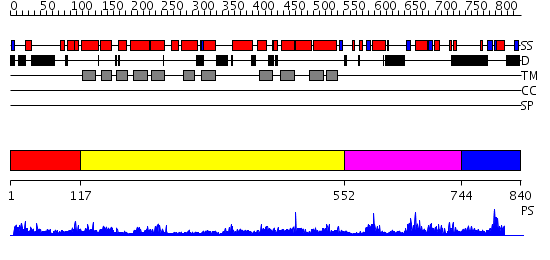
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 1.192989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [117..551] | 0.988 | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | |
| 3 | View Details | [552..743] | 1.51 | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | |
| 4 | View Details | [744..840] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|