













| General Information: |
|
| Name(s) found: |
PUR6_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 552 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEKQVVGIL GGGQLGRMMV EAAHRLNIKC IILDAANSPA KQIDGGREHI DASFTDPDAI 60
61 VELSKKCTLL TTEIEHINTD ALAAVTKSVA VEPSPATLRC IQDKYLQKQH LQVFKIALPE 120
121 FCDAPDQESV EKAGQEFGYP FVLKSKTLAY DGRGNYVVHQ PSEIPTAIKA LGDRPLYVEK 180
181 FVPFSMEIAV MVVRSLDGKV YAYPTTETIQ KDNVCHLVYA PARLPFSIQQ RAQTLAMDAV 240
241 RTFEGAGIYG VEMFVLQDGE TILLNEIAPR PHNSGHYTIE ACPTSQFEAH LRAICGLPFS 300
301 EINTQLSTST THALMVNILG TDDPDYVSKI AKRSLSIPGA TLHLYGKAES RKGRKMGHVT 360
361 IISDSPQECE RRYQMLLDVK DPVESPVVGI IMGSDSDLSK MKDAAVILDE FKVPYELTIV 420
421 SAHRTPDRMV TYARTAASRG LRVIIAGAGG AAHLPGMVAA MTPLPVIGVP VKGSTLDGVD 480
481 SLHSIVQMPR GVPVATVAIN NSQNAGILAC RILATFQPSL LAAMESFMDN MKEIVLEKAD 540
541 KLEKVGWKNY SA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
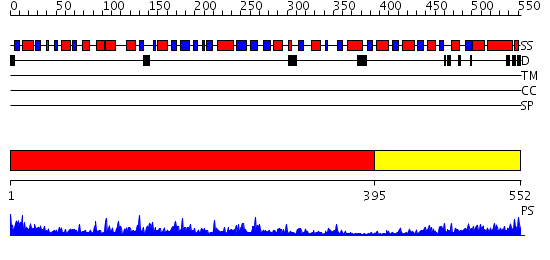
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..394] | 57.0 | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3 | |
| 2 | View Details | [395..552] | 40.69897 | N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.60 |
Source: Reynolds et al. (2008)