













| General Information: |
|
| Name(s) found: |
FBpp0300846
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1939 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKCIPIFKG CNRQVEFVDK RHCSLPQVPE EILRYSRTLE ELFLDANHIR DLPKNFFRLH 60
61 RLRKLGLSDN EIGRLPPDIQ NFENLVELDV SRNDIPDIPD DIKHLQSLQV ADFSSNPIPK 120
121 LPSGFSQLKN LTVLGLNDMS LTTLPADFGS LTQLESLELR ENLLKHLPET ISQLTKLKRL 180
181 DLGDNEIEDL PPYLGYLPGL HELWLDHNQL QRLPPELGLL TKLTYLDVSE NRLEELPNEI 240
241 SGLVSLTDLD LAQNLLEALP DGIAKLSRLT ILKLDQNRLQ RLNDTLGNCE NMQELILTEN 300
301 FLSELPASIG QMTKLNNLNV DRNALEYLPL EIGQCANLGV LSLRDNKLKK LPPELGNCTV 360
361 LHVLDVSGNQ LLYLPYSLVN LQLKAVWLSE NQSQPLLTFQ PDTDAETGEQ VLSCYLLPQQ 420
421 EYQPITPARD LESDSEPFEE REPSRTVVKF SEEATQEKET PFVRQNTPHP KDLKAKAQKL 480
481 KVERSRNEEH ANLVTLPEEN GTKLAETPTE TRTIANNHQQ QPHPVQQPIV GVNSKQPVVV 540
541 GVVTPTTTTV APTGVQGGSE GASSTANNVK AATAAVVAEL AATVGGSDEV QDDDEQEDEF 600
601 ESDRRVGFQV EGEDDDFYKR PPKLHRRDTP HHLKNKRVQH LTDKQASEIL ANALASQERN 660
661 DTTPQHSLSG KVTSPIEEEE QLEVEQEQQQ QQQQHPFDSS LSPISAGKTA EASTDPANQA 720
721 MFNVKIKVKE GNNTLPRNGQ RSNGNFSYSG AGSGRNSPAM SSLRSSLASS NRSPSGSLQR 780
781 KGKRVRIVTS YDPIDRDYER QRQHYDNLDG VTELRLEQYE IHIERTAAGL GLSIAGGKGS 840
841 TPFKGDDDGI FISRVTEAGP ADLAGLKVGD KVIKVNGIVV VDADHYQAVQ VLKACGAVLV 900
901 LVVQREVTRL IGHPVFSEDG SVSQISVETR PLVADAPPAA SISHERYIPA PIEIVPQQQH 960
961 LQQQQQQPIQ QVAPTHSYSG NVFATPTAAQ TVQPAVSAAP NGLLLNGREA PLSYIQLHTT1020
1021 LIRDQIGQGL GFSIAGGKGS PPFKDDCDGI FISRITEGGL AYRDGKIMVG DRVMAINGND1080
1081 MTEAHHDAAV ACLTEPQRFV RLVLQREYRG PLEPPTSPRS PAVLNSLSPS GYLANRPANF1140
1141 SRSVVEVEQP YKYNTLATTT PTPKPTVPAS ISNNNNTLPS SKTNGFATAA AATIDSSTGQ1200
1201 PVPAPRRTNS VPMGDGDIGA GSTTSGDSGE AQPSSLRPLT SDDFQAMIPA HFLSGGSQHQ1260
1261 VHVARPNEVG VSAVTVNVNK PQPDLPMFPA APTELGRVTE TITKSTFTET VMTRITDNQL1320
1321 AEPLISEVVL PKNQGSLGFS IIGGTDHSCV PFGTREPGIF ISHIVPGGIA SKCGKLRMGD1380
1381 RILKVNEADV SKATHQDAVL ELLKPGDEIK LTIQHDPLPP GFQEVLLSKA EGERLGMHIK1440
1441 GGLNGQRGNP ADPSDEGVFV SKINSVGAAR RDGRLKVGMR LLEVNGHSLL GASHQDAVNV1500
1501 LRNAGNEIQL VVCKGYDKSN LIHSIGQAGG MSTGFNSSAS CSGGSRQGSR ASETGSELSQ1560
1561 SQSVSSLDHE EDERLRQDFD VFASQKPDAQ QPTGPSVLAA AAAMVHGASS PTPPAATSNI1620
1621 TPLPTAAAVA SADLTAPDTP ATQTVALIHA EQQAHQQQQQ TQLAPLGQEK STQEKVLEIV1680
1681 RAADAFTTVP PKSPSEHHEQ DKIQKTTTVV ISKHTLDTNP TTPTTPAAPL SIAGAESANS1740
1741 AGAPSPAVPA STPGSAPVLP AVAVQTQTQT TSTEKDEEEE SQLQSTPASR DGAEEQQEEV1800
1801 RAKPTPTKVP KSVSDKKRFF ESAMEDQHKP TQKTDKVFSF LSKDEVEKLR QEEERKIATL1860
1861 RRDKNSRLLD AANDNIDKDA AQQRTKSNSN SSSGDDNDDS DQEEGIARGD SVDNAALGHF1920
1921 DDAEDMRNPL DEIEAVFRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
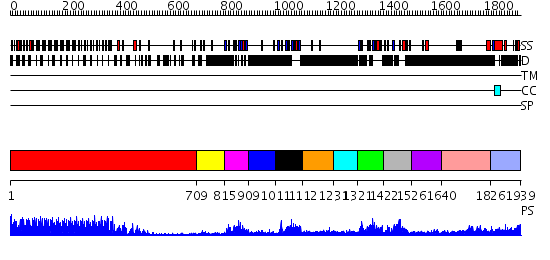
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..708] | 62.522879 | The molecular structure of toll-like receptor 3 ligand binding domain | |
| 2 | View Details | [709..814] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [815..908] | 22.045757 | No description for 2g2lA was found. | |
| 4 | View Details | [909..1010] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1011..1111] | 22.154902 | Solution structure of the first PDZ domain of scribble homolog protein (hScrib) | |
| 6 | View Details | [1112..1230] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1231..1320] | 26.69897 | Structural Insights into the Inter-domain Chaperoning of Tandem PDZ Domains in Glutamate Receptor Interacting Proteins | |
| 8 | View Details | [1321..1421] | 4.78 | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | |
| 9 | View Details | [1422..1525] | 24.0 | Solution structure of the fourth PDZ domain of human scribble (KIAA0147 protein) | |
| 10 | View Details | [1526..1639] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1640..1825] | 2.134995 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1826..1939] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 |
|
|||||||||
| 6 | No functions predicted. | |||||||||
| 7 |
|
|||||||||
| 8 |
|
|||||||||
| 9 |
|
|||||||||
| 10 | No functions predicted. | |||||||||
| 11 | No functions predicted. | |||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)