













| General Information: |
|
| Name(s) found: |
mld-PB /
FBpp0110312
[FlyBase]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1728 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
ecdysone biosynthetic process
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTMHIKRPF ICTLHIRHMY LCMFLPCINR DDGLPQEICR QCMAQFLMVA KFRRKCVRMQ 60
61 QRLQDFAQEI SDRQTARQAK RQQKVQKKTH DRWLQLEPTS HEEQQPIRKR RMVSESDGDS 120
121 TAAAAAAEAA TATATTPAKQ VRTDFESLPP PKPSPQQHRV RKYRTLSIDC EMERPAASIN 180
181 ASSMPWKTKA ARKELSENWA SSCSTAVMSP NRQRSSTSSS EHMSDLSVGP CCGFESDSNP 240
241 SSTSNSARRT VILVKQQSPE KTSPMYDLMP RRGRRPRKAP IYMPAKRQRR SLVRKEPLYR 300
301 KAQPVNEENK KSDQKDIYPK ESRMEGLAPS CDEAIDSTSD APMEVASNVV QKKCDEICLN 360
361 GLIDSRNQLG AMEMEPVNEM IASNGVDKEL TLSVQNNKET TSEEQIFEAT SVIKESVQPE 420
421 QSAEDIQVRT EIESIANDIR AEADLVQSEE LTSSTETSES QPKAVEQLES MGKQESYALP 480
481 QNMEAHESVQ QTQTVDESDS ADQREIIVSI PLEVLDENLQ RRLCVDPETE AANRQSADPK 540
541 EFALRPDDVN NNENDENFCQ SATADSFNSA VALRTNKIEA TPPGGTDSDD TDTDEVPIVP 600
601 SGPTPMDFMA EFAKHCANGI EQLKVTTPVP SPTPSDLVPL NDLEAKQKLE VEMNDVETTL 660
661 NGILNEMQDQ HMYTPNCAHI DEFLTPADYA TLTEQESSVS SPPNPFEQSS VAEPKDEKPS 720
721 ATEDPFDNSN FTNELIGFQN DIPCFENIET PEEGQNLHLE LTQFLRDLQP SQSDPPAGEQ 780
781 VEKIEMETQP STVQMDVQSN LPVQAADTQP QNESANHSPF ESPISSPQVQ SQIELQMEPQ 840
841 IETQMQPQIE TRMQHDVLSV QSPQNHQPQV QSHDQSQMQH QIPPQIEPQI EVQNQEQNTL 900
901 QLVPQAQHQV VPHVQLQSQL VPHVQTQGTT TTVTYEQIYQ QHIVQQTVQQ SSNKMQVISL 960
961 PSAGSETTRQ WQTQQSQQQQ QVQWSSVGGL EAIKSAPVES VAPTTTTYYI SASDLYPQQT1020
1021 ETYTMLQPAS NESYVIEQVN HQQQHCQQQI QMSQQQHQRQ HQQQQAQQQQ QPIMILIQQP1080
1081 EIQQPVASAS NPQQAPLHIY NAATSNDVHQ MQHTQRQQIR QQYQHQQYQR LQQPQQTRVV1140
1141 QSHPVLGMPG ATSISTKVTP IPVTAARGRP LTLKCRFCHN GPRFSSSLEY SRHIIDLHPA1200
1201 VAPFNCPHCP MAFAGRTKRS QHILSHHVVQ QYQCGQCSHV FPAQKALDVH IQRFHMTLKT1260
1261 DPSGAVKVED VQLQITSERR PRGRPYKPRL QLQQHQLQPQ QKLQVQQHQQ QQKALQQLPQ1320
1321 AQHHQQQQQP QLQQEVLQQV QQLQVQVQPQ QHQQHQQVLQ VQQPQQQPLM QPQSPHPSTV1380
1381 EMQASPTTPR KILCCPDCED CTSGHSHANE QFEELQTLQA PPTVLTPPST IVSVPSPQPM1440
1441 VYSQHITMPS PEQSEPDSTT TLRQYRKRGV IVGPQGPLHL ATPVASPSPS SSPSSSTVDH1500
1501 IPPASPATPA SPAPPPSPAV ASTVQVSELR TSHHCLYCEE RFTNEISLKK HHQLAHGALT1560
1561 TMPYVCTICK RGYRMRTALH RHMESHDVEG RPYECNICRV RFPRPSQLTL HKITVHLLSK1620
1621 PHTCDECGKQ FGTESALKTH IKFHGAHMKT HLPLGVFRNE DAASKSPSNN NSHASSDKLE1680
1681 NPATPIEETP LTPMSSGGHL YHSPDEYPNS VESCAGNSVA LESYATTP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
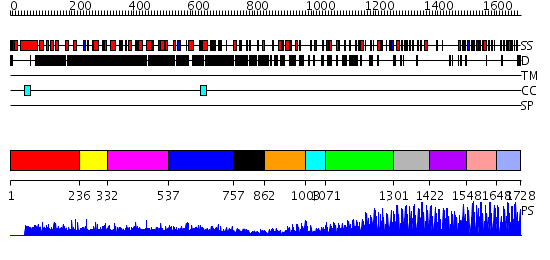
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..235] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [236..331] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [332..536] | 1.025979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [537..756] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [757..861] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [862..1002] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1003..1070] | 1.362993 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1071..1300] | 3.37 | No description for 2i13A was found. | |
| 9 | View Details | [1301..1421] | 24.522879 | Ying-yang 1 (yy1, zinc finger domain) | |
| 10 | View Details | [1422..1547] | 62.154902 | No description for 2i13A was found. | |
| 11 | View Details | [1548..1647] | 23.522879 | Five-finger GLI1 | |
| 12 | View Details | [1648..1728] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. | ||||||
| 9 | No functions predicted. | ||||||
| 10 | No functions predicted. | ||||||
| 11 |
|
||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)