













| General Information: |
|
| Name(s) found: |
p120ctn-PB /
FBpp0110405
[FlyBase]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: |
adherens junction
[IDA][IPI][ISS]
centrosome [IDA] cytoplasm [IDA] |
| Biological Process: |
compound eye morphogenesis
[IGI][IMP]
cell adhesion [IGI] adherens junction maintenance [IMP] |
| Molecular Function: |
binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSTHINMDL SLTHEPANMQ SQFGSFQNYD QFSAAGYSST PLYITKPTGV GAITKVAPHC 60
61 SQNKIETNST DNSIPDIENH PLATTVRYVQ AGQYEDTSEY GVAGLTCGID SEYTAEAVVP 120
121 YCYTSDANYT GIQNTTSSIK PFSGRPFNDN INGAEAVADS QCRTFKSSAE FKLPQFAMSS 180
181 INTVPQRLEE KDDYIEGSEN DICSTMRWRD PNLSEVISFL SNPSSAIKAN AAAYLQHLCY 240
241 MDDPNKQRTR SLGGIPPLVR LLSYDSPEIH KNACGALRNL SYGRQNDENK RGIKNAGGIA 300
301 ALVHLLCRSQ ETEVKELVTG VLWNMSSCED LKRSIIDEAL VAVVCSVIKP HSGWDAVCCG 360
361 ETCFSTVFRN ASGVLRNVSS AGEHARGCLR NCEHLVECLL YVVRTSIEKN NIGNKTVENC 420
421 VCILRNLSYR CQEVDDPNYD KHPFITPERV IPSSSKGENL GCFGTNKKKK EANNSDALNE 480
481 YNISADYSKS SVTYNKLNKG YEQLWQPEVV QYYLSLLQSC SNPETLEAAA GAIQNLSACY 540
541 WQPSIDIRAT VRKEKGLPIL VELLRMEVDR VVCAVATALR NLAIDQRNKE LIGKYAMRDL 600
601 VQKLPSGNVQ HDQNTSDDTI TAVLATINEV IKKNPEFSRS LLDSGGIDRL MNITKRKEKY 660
661 TSCVLKFASQ VLYTMWQHNE LRDVYKKNGW KEQDFVSKHF TAHNTPPSSP NNVNNTLNRP 720
721 MASQGRTRYE DRTIQRGTST LYSANDSSGA VMSNESAMLS EMVRKQL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
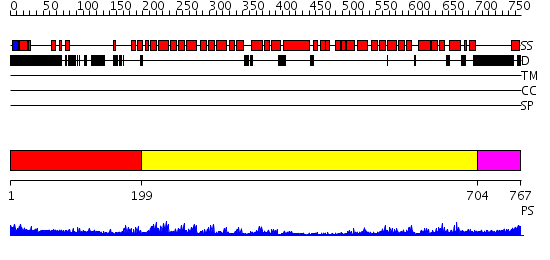
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | 40.0 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 2 | View Details | [199..703] | 60.221849 | Structure of the armadillo repeat domain of plakophilin 1 | |
| 3 | View Details | [704..767] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)