













| General Information: |
|
| Name(s) found: |
baz-PB /
FBpp0110299
[FlyBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1520 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[TAS]
adherens junction [IEP] subapical complex [TAS] apical part of cell [NAS] spot adherens junction [IDA] cell-cell adherens junction [NAS] cytoplasm [NAS] cell cortex [TAS] apical junction complex [IEP] apical cortex [NAS][TAS] apical plasma membrane [NAS][TAS] |
| Biological Process: |
establishment of apical/basal cell polarity
[IMP]
germarium-derived oocyte fate determination [IMP] protein localization [TAS] germ-band extension [IMP][TAS] border follicle cell delamination [IMP] cytokinesis [IMP] border follicle cell migration [IMP] ovarian follicle cell-cell adhesion [IMP] asymmetric protein localization [TAS] establishment or maintenance of neuroblast polarity [NAS] oocyte axis specification [TAS] basal protein localization [TAS] establishment or maintenance of epithelial cell apical/basal polarity [IMP][NAS][TAS] apical protein localization [NAS] establishment or maintenance of polarity of embryonic epithelium [NAS] establishment of mitotic spindle localization [TAS] cell-cell junction assembly [NAS] establishment or maintenance of cell polarity [NAS] asymmetric cell division [TAS] synapse assembly [IMP] morphogenesis of an epithelium [NAS][TAS] asymmetric protein localization involved in cell fate determination [IMP] zonula adherens assembly [IMP][NAS][TAS] morphogenesis of a polarized epithelium [TAS] asymmetric neuroblast division [IGI][IMP] |
| Molecular Function: |
protein kinase C binding
[ISS]
protein binding [IPI][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFDVPPKCPA LANKLGGLFG WRHTYKVAAK QEGIPHGQLH KSYSLTLPKR PPSPSTYSCV 60
61 KPDSWVTVTH LQTQSGILDP DDCVRDVADD REQILAHFDD PGPDPGVPQG GGDGASGSSS 120
121 VGTGSPDIFR DPTNTEAPTC PRDLSTPHIE VTSTTSGPMA GLGVGLMVRR SSDPNLLASL 180
181 KAEGSNKRWS AAAPHYAGGD SPERLFLDKA GGQLSPQWEE DDDPSHQLKE QLLHQQQPHA 240
241 ANGGSSSGNH QPFARSGRLS MQFLGDGNGY KWMEAAEKLQ NQPPAQQTYQ QGSHHAGHGQ 300
301 NGAYSSKSLP RESKRKEPLG QAYESIREKD GEMLLIINEY GSPLGLTALP DKEHGGGLLV 360
361 QHVEPGSRAE RGRLRRDDRI LEINGIKLIG LTESQVQEQL RRALESSELR VRVLRGDRNR 420
421 RQQRDSKVAE MVEVATVSPT RKPHAAPVGT SLQVANTRKL GRKIEIMLKK GPNGLGFSVT 480
481 TRDNPAGGHC PIYIKNILPR GAAIEDGRLK PGDRLLEVDG TPMTGKTQTD VVAILRGMPA 540
541 GATVRIVVSR QQELAEQADQ PAEKSAGVAV APSVAPPAVP AAAAPAPPIP VQKSSSARSL 600
601 FTHQQQSQLN ESQHFIDAGS ESAASNDSLP PSSNSWHSRE ELTLHIPVHD TEKAGLGVSV 660
661 KGKTCSNLNA SGSSASSGSN GLMKHDGDLG IFVKNVIHGG AASRDGRLRM NDQLLSVNGV 720
721 SLRGQNNAEA METLRRAMVN TPGKHPGTIT LLVGRKILRS ASSSDILDHS NSHSHSHSNS 780
781 SGGSNSNGSG NNNNSSSNAS DNSGATVIYL SPEKREQRCN GGGGGGSAGN EMNRWSNPVL 840
841 DRLTGGICSS NSAQPSSQQS HQQQPHPSQQ QQQQRRLPAA PVCSSAALRN ESYYMATNDN 900
901 WSPAQMHLMT AHGNTALLIE DDAEPMSPTL PARPHDGQHC NTSSANPSQN LAVGNQGPPI 960
961 NTVPGTPSTS SNFDATYSSQ LSLETNSGVE HFSRDALGRR SISEKHHAAL DARETGTYQR1020
1021 NKKLREERER ERRIQLTKSA VYGGSIESLT ARIASANAQF SGYKHAKTAS SIEQRETQQQ1080
1081 LAAAEAEARD QLGDLGPSLG MKKSSSLESL QTMVQELQMS DEPRGHQALR APRGRGREDS1140
1141 LRAAVVSEPD ASKPRKTWLL EDGDHEGGFA SQRNGPFQSS LNDGKHGCKS SRAKKPSILR1200
1201 GIGHMFRFGK NRKDGVVPVD NYAVNISPPT SVVSTATSPQ LQQQQQQQLQ QHQQQQQIPT1260
1261 AALAALERNG KPPAYQPPPP LPAPNGVGSN GIHQNDIFNH RYQHYANYED LHQQHQQHQI1320
1321 SGGDSTTSIS ETLSESTLEC MRQQVIRQRI KVEAESRRHQ HYHSQRSARS QDVSMHSTSS1380
1381 GSQPGSLAQP QAQSNGVRPM SSYYEYETVQ QQRVGSIKHS HSSSATSSSS SPINVPHWKA1440
1441 AAMNGYSPAS LNSSARSRGP FVTQVTIREQ SSGGIPAHLL QQHQQQQLQQ QQPTYQTVQK1500
1501 MSGPSQYGSA AGSQPHASKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
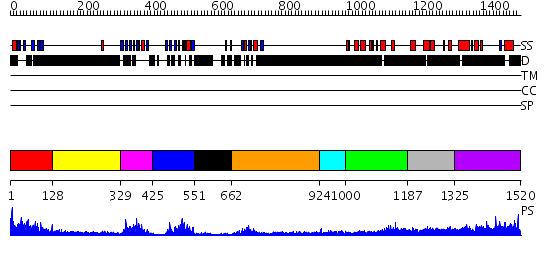
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 18.69897 | No description for 2dazA was found. | |
| 2 | View Details | [128..328] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [329..424] | 19.69897 | Synaptic protein PSD-95 | |
| 4 | View Details | [425..550] | 25.0 | The crystal structure of the 13th PDZ domain of MPDZ | |
| 5 | View Details | [551..661] | 3.35 | No description for 2iwqA was found. | |
| 6 | View Details | [662..923] | 3.97 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 7 | View Details | [924..999] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1000..1186] | 32.09691 | Psd-95; Guanylate kinase-like domain of Psd-95 | |
| 9 | View Details | [1187..1324] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1325..1520] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)