













| General Information: |
|
| Name(s) found: |
RASAL2
[HGNC (HUGO)]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1139 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQTPEVPAER SPRRRSISGT STSEKPNSMD TANTSPFKVP GFFSKRLKGS IKRTKSQSKL 60
61 DRNTSFRLPS LRSTDDRSRG LPKLKESRSH ESLLSPCSTV ECLDLGRGEP VSVKPLHSSI 120
121 LGQDFCFEVT YLSGSKCFSC NSASERDKWM ENLRRTVQPN KDNCRRAENV LRLWIIEAKD 180
181 LAPKKKYFCE LCLDDTLFAR TTSKTKADNI FWGEHFEFFS LPPLHSITVH IYKDVEKKKK 240
241 KDKNNYVGLV NIPTASVTGR QFVEKWYPVS TPTPNKGKTG GPSIRIKSRF QTITILPMEQ 300
301 YKEFAEFVTS NYTMLCSVLE PVISVRNKEE LACALVHILQ STGRAKDFLT DLVMSEVDRC 360
361 GEHDVLIFRE NTIATKSIEE YLKLVGQQYL HDALGEFIKA LYESDENCEV DPSKCSSSEL 420
421 IDHQSNLKMC CELAFCKIIN SYCVFPRELK EVFASWKQQC LNRGKQDISE RLISASLFLR 480
481 FLCPAIMSPS LFNLMQEYPD DRTSRTLTLI AKVIQNLANF AKFGNKEEYM AFMNDFLEHE 540
541 WGGMKRFLLE ISNPDTISNT PGFDGYIDLG RELSVLHSLL WEVVSQLDKG ENSFLQATVA 600
601 KLGPLPRVLA DITKSLTNPT PIQQQLRRFT EHNSSPNVSG SLSSGLQKIF EDPTDSDLHK 660
661 LKSPSQDNTD SYFRGKTLLL VQQASSQSMT YSEKDERESS LPNGRSVSLM DLQDTHAAQV 720
721 EHASVMLDVP IRLTGSQLSI TQVASIKQLR ETQSTPQSAP QVRRPLHPAL NQPGGLQPLS 780
781 FQNPVYHLNN PIPAMPKASI DSSLENLSTA SSRSQSNSED FKLSGPSNSS MEDFTKRSTQ 840
841 SEDFSRRHTV PDRHIPLALP RQNSTGQAQI RKVDQGGLGA RAKAPPSLPH SASLRSTGSM 900
901 SVVSAALVAE PVQNGSRSRQ QSSSSRESPV PKVRAIQRQQ TQQVQSPVDS ATMSPVERTA 960
961 AWVLNNGQYE EDVEETEQNL DEAKHAEKYE QEITKLKERL RVSSRRLEEY ERRLLVQEQQ1020
1021 MQKLLLEYKA RLEDSEERLR RQQEEKDSQM KSIISRLMAV EEELKKDHAE MQAVIDAKQK1080
1081 IIDAQEKRIV SLDSANTRLM SALTQVKERY SMQVRNGISP TNPTKLSITE NGEFKNSSC |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
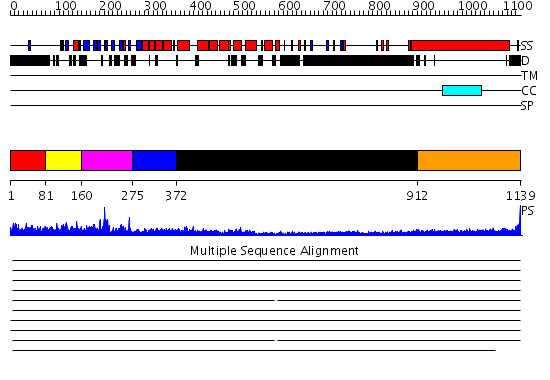
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..159] | 3.522879 | GEF of intersectin | |
| 3 | View Details | [160..274] | 20.522879 | No description for 2enpA was found. | |
| 4 | View Details | [275..371] | 28.69897 | RAS-GTPASE-ACTIVATING DOMAIN OF HUMAN P120GAP | |
| 5 | View Details | [372..911] | 22.221849 | Heavy meromyosin subfragment | |
| 6 | View Details | [912..1139] | 6.154902 | Tropomyosin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)