













| General Information: |
|
| Name(s) found: |
AKAP8
[HGNC (HUGO)]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 692 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQGYGGYGA WSAGPANTQG AYGTGVASWQ GYENYNYYGA QNTSVTTGAT YSYGPASWEA 60
61 AKANDGGLAA GAPAMHMASY GPEPCTDNSD SLIAKINQRL DMMSKEGGRG GSGGGGEGIQ 120
121 DRESSFRFQP FESYDSRPCL PEHNPYRPSY SYDYEFDLGS DRNGSFGGQY SECRDPARER 180
181 GSLDGFMRGR GQGRFQDRSN PGTFMRSDPF VPPAASSEPL STPWNELNYV GGRGLGGPSP 240
241 SRPPPSLFSQ SMAPDYGVMG MQGAGGYDST MPYGCGRSQP RMRDRDRPKR RGFDRFGPDG 300
301 TGRKRKQFQL YEEPDTKLAR VDSEGDFSEN DDAAGDFRSG DEEFKGEDEL CDSGRQRGEK 360
361 EDEDEDVKKR REKQRRRDRT RDRAADRIQF ACSVCKFRSF DDEEIQKHLQ SKFHKETLRF 420
421 ISTKLPDKTV EFLQEYIVNR NKKIEKRRQE LMEKETAKPK PDPFKGIGQE HFFKKIEAAH 480
481 CLACDMLIPA QPQLLQRHLH SVDHNHNRRL AAEQFKKTSL HVAKSVLNNR HIVKMLEKYL 540
541 KGEDPFTSET VDPEMEGDDN LGGEDKKETP EEVAADVLAE VITAAVRAVD GEGAPAPESS 600
601 GEPAEDEGPT DTAEAGSDPQ AEQLLEEQVP CGTAHEKGVP KARSEAAEAG NGAETMAAEA 660
661 ESAQTRVAPA PAAADAEVEQ TDAESKDAVP TE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
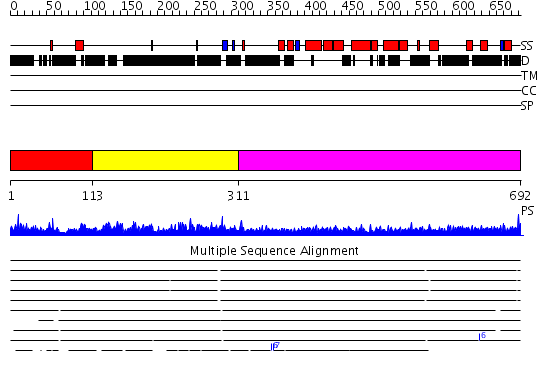
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [113..310] | 9.39794 | No description for PF04988.3 was found. No confident structure predictions are available. | |
| 3 | View Details | [311..692] | 8.0 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)