













| General Information: |
|
| Name(s) found: |
gi|108862892
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 1041 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGKYLWKPL LRFSGESYRF SPLSRSCAQD RRGDGVRPRQ IRRLRAGEAG SRAVSGGNRA 60
61 VRDGNRLGRR ARGNPSVLKV FARVEMSDSR DPVWEHGENI PPGWRCKYCH TKRGGGGATR 120
121 LKQHLAARGK GVTYCNSVPP DVREFFCREL DRIKDAGDQR KSDSGRRVEA ARVNYYDLTG 180
181 DADEEEQMEA AIAVSRQDEN FRRDVEERGG TYEHGGGSGS AQPEARKGRS NPITNMLRRA 240
241 TSHRESPAVR DYNLASAKAP VQPRIDTGFF TKKGKQARQA IGESWARFFF TAGIPGRNAD 300
301 NPYFVSAVRE TQKWGESVPS PTGNEIDGKY LDSTEKDVKK QFDRFKKDWD EYGVTIMCDS 360
361 WTGPTSMSVI NFLIYCNGIM FFHKSIDATG QSQDANFVLK EIRKVVREIG SEHVVQIITY 420
421 NGSNYKKACR LLRQEYKTIV WQPCVAHTVN LMLKEVGKMP DHEMVIESAR KICRWLYNHN 480
481 KLHAMMVLAI GGELVKWNAT RFGTNYMFLQ SFLRKRDLFM QWMASSVFMQ SKFSGTLEGK 540
541 YAHACLSSLS WWENLEAVVN SVQPMYSFLR FADEDKNPNL SEVLLRYQLL KMEYDSLFAN 600
601 QRDKFEAYME IVNRRMHNLT NETLINAAAA LNPRTHYAYS PSATVFQDLR QAFEWMTDID 660
661 TAAAALLEVE MYRRKTGEFG RALARRMAID GKTSPAQWWS MFASDTPNLK KLALRLVGQC 720
721 CSSSGCERNW STFAFVHTKV RNRLTHKKLN KLVYVNYNLR LRIQQANAQI RVEDDDPLQR 780
781 LADLSFYETN NHISAWMDNA RSNACPELDE DSAESDAPLP SQLVSDLVNL DDLRRTTGAS 840
841 SIAEWADTNV GDTHIGKRKT RKPPKARPSK KVKGKGPRST SVDSDEETQG SPEYQESNDS 900
901 SSRTETDDGD DDGEGDFTHA TQDQDHGAPS SQRTTIAPGV RQQQQFSDIQ QDSSSSFSAS 960
961 SFESGYPTYV YNRPQASDYP ATTWVYEWQE PQCSNISRLP SSYRGYRTKP CGYRVHTVDF1020
1021 EFKNLETKFV RFFAVTARLP R |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
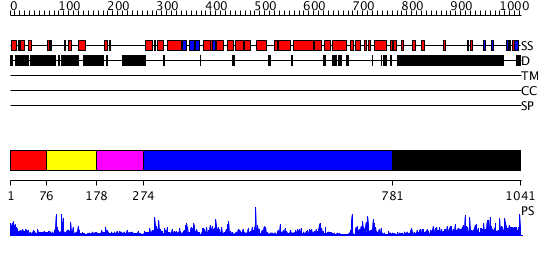
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..177] | 18.886057 | No description for PF07567.4 was found. No confident structure predictions are available. | |
| 3 | View Details | [178..273] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [274..780] | 52.0 | Three-dimensional structure of the Hermes DNA transposase | |
| 5 | View Details | [781..1041] | 10.206997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)