













| General Information: |
|
| Name(s) found: |
gi|115451109
[NCBI NR]
gi|113547626 [NCBI NR] gi|108706494 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 870 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPAAASPDD RIRSYEDFAR VHAYLLAASG IPPSLHQRLY RKLADEVFDG GEAFSVEPCE 60
61 GGRQRRLVLA AEWTLGRESD VFLVDHAWSF RLSDALKQLR EVPGLAERMA ALMCVDLDER 120
121 TELEEADEQD NGNGGSLESA LEVVEKERTR IQEKGSDFAA WLELEELGID DDMLIALDLS 180
181 SKFPNMVALN LWGNKLQDPE KIMKGIGECR RLKALWLNEN PALKEGVDKV ILDGLPELEI 240
241 YNSHFTRKAG EWALGFCGDI IGADNPCSSA ESIPLENIAS LDLSDRCIHK LPVVFSPRKL 300
301 SSLLSLNIRG NPLDQMSSDD LLKLISGFTQ LQELEVDIPG SLGNSAISIL ECLPNLSLLN 360
361 GINVASIIES GKHIIDSALK PRLPEWSPQE SLPERVIGAM WLYLMTYRLA DEEKIDETPV 420
421 WYVMDELGSA MRHSDDANFR IAPFLFMPDG KLASAISYTI LWPVHDVHTG EECTRDFLFG 480
481 VGEDKQRSAR LTAWFHTPEN YFIQEFRKYK EQLQSSSICP SRKVTPVTKS IRPSDGHALR 540
541 VFTDIPQVEE FLTRPEFVLT SDPKEADIIW VSMQVDSELK NALGLTDQQY TNQFPFEACL 600
601 VMKHHLAETI HKAWGSPEWL QPTYNLETHL SPLIGDYCVR KRDGMDNLWI MKPWNMARTI 660
661 DTTVTGDLSA IIRLMETGPK ICQKYIECPA LFQGRKFDLR YIVFVRSICP LEIFLSDVFW 720
721 VRLANNQYTL EKTSFFEYET HFTVMNYIGR MNHMNTPEFV KEFEKEHQVK WLEIHGRIRD 780
781 MIRCVFESAT AVHPEMQNPF SRAIYGVDVM LDNKFNPKIL EVTYCPDCTR ACKYDTQALV 840
841 GSQGVIRGTE FFNTVFGCLF LDELKDVSPL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
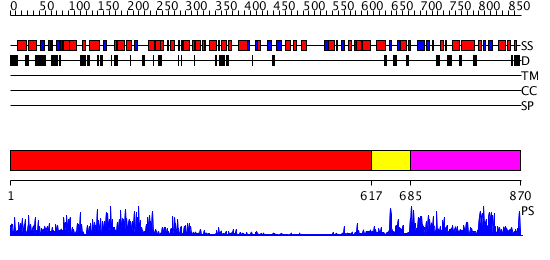
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..616] | 14.09691 | No description for 2z63A was found. | |
| 2 | View Details | [617..684] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [685..870] | 8.198976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)