













| General Information: |
|
| Name(s) found: |
gi|115450235
[NCBI NR]
gi|113547189 [NCBI NR] gi|108705795 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 597 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDWASDSDN DKFEWDTDGE AETSSAPALR NIDAPGPSTR LPQDANGKAN GSGALVAEFM 60
61 GMGFPKEMIL KAIKEIGDTD TEQLLELLLT YQAIGGDASV GNCSASACAP QTLEVDEEED 120
121 DTNWDEYDTA GNCDRTPHSD GSGDEDFFQE MSEKDEKMKS LVNMGFPEDE AKMAIDRCLD 180
181 APVAVLVDSI YASQEAGNGY SANLSDYEDT EFSSFGGRKK TRFVDGSKKR KRYGSGPSGN 240
241 QVPFDGSHEE PMPLPNPMVG FSLPNERLRS VHRNLPDQAL GPPFFYYENV ALAPKGVWTT 300
301 ISRFLYDIQP EFVDSKYFCA AARKRGYIHN LPIENRSPVL PMPPKTISEA FPNTKRWWPS 360
361 WDPRRQFNCL QTCMASAKLT ERIRCALGRF SDVPTPQVQK YVLDECRKWN LVWVGKNKVA 420
421 PLEPDEMEFL LGYPRNHTRG VSRTERYRAL GNSFQVDTVA YHLSVLRDLF PNGMNVLSLF 480
481 SGIGGAEVAL HRLGIHMKTV ISVEKSEVNR TILKSWWDQT QTGTLIEIAD VRHLTTERIE 540
541 TFIRRFGGFD LVIGGSPCNN LAGSNRHHRD GLEGEHSALF YDYIRILEHV KATMSAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
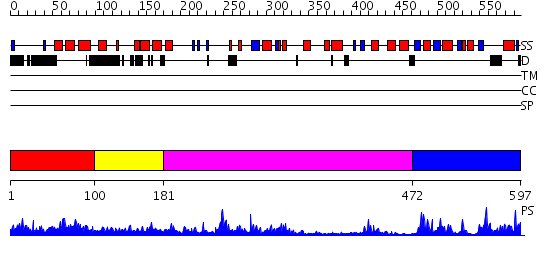
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | 1.16 | Solution Structure of UBA domain of Human Ubiquitin Associated Protein 1 (UBAP1) | |
| 2 | View Details | [100..180] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [181..471] | 44.522879 | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions | |
| 4 | View Details | [472..597] | 4.045757 | crystal structure of a putative RNA methyltransferase PH1948 from Pyrococcus horikoshii |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 |
|
|||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)