













| General Information: |
|
| Name(s) found: |
MON1A
[HGNC (HUGO)]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Homo sapiens |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATDMQRKRS SECLDGTLTP SDGQSMERAE SPTPGMAQGM EPGAGQEGAM FVHARSYEDL 60
61 TESEDGAASG DSHKEGTRGP PPLPTDMRQI SQDFSELSTQ LTGVARDLQE EMLPGSSEDW 120
121 LEPPGAVGRP ATEPPREGTT EGDEEDATEA WRLHQKHVFV LSEAGKPVYS RYGSEEALSS 180
181 TMGVMVALVS FLEADKNAIR SIHADGYKVV FVRRSPLVLV AVARTRQSAQ ELAQELLYIY 240
241 YQILSLLTGA QLSHIFQQKQ NYDLRRLLSG SERITDNLLQ LMARDPSFLM GAARCLPLAA 300
301 AVRDTVSASL QQARARSLVF SILLARNQLV ALVRRKDQFL HPIDLHLLFN LISSSSSFRE 360
361 GEAWTPVCLP KFNAAGFFHA HISYLEPDTD LCLLLVSTDR EDFFAVSDCR RRFQERLRKR 420
421 GAHLALREAL RTPYYSVAQV GIPDLRHFLY KSKSSGLFTS PEIEAPYTSE EEQERLLGLY 480
481 QYLHSRAHNA SRPLKTIYYT GPNENLLAWV TGAFELYMCY SPLGTKASAV SAIHKLMRWI 540
541 RKEEDRLFIL TPLTY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
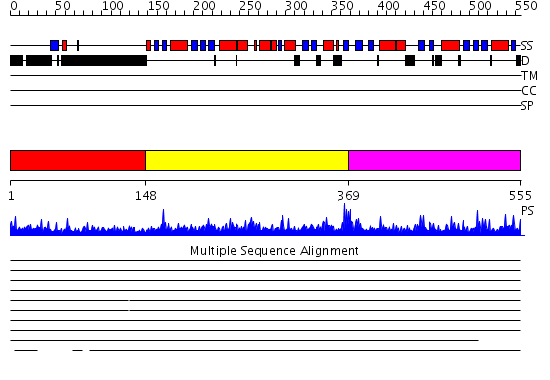
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 4.0 | Structure of the DNA repair protein hHR23a | |
| 2 | View Details | [148..368] | 1.11 | AP1 clathrin adaptor core | |
| 3 | View Details | [369..555] | 3.052956 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)