













| General Information: |
|
| Name(s) found: |
gi|50346005
[NCBI NR]
gi|13623609 [NCBI NR] |
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCNQGRGRES SRGGLHVQGS CRGLSRSPQQ ETGFAKAPAG TDCFFHCSPG SRGQGDRKEE 60
61 VTSEPGGTSI MSRLGGMNAA VRAVTRMGIY VGAKVFLIYE GYEGLVEGGE NIKQANWLSV 120
121 SNIIQLGGTI IGSARCKAFT TREGRRAAAY NLVQHGITNL CVIGGDGSLT GANIFRSEWG 180
181 SLLEELVAEG KISETTARTY SHLNIAGLVG SIDNDFCGTD MTIGTDSALH RIMEVIDAIT 240
241 TTAQSHQRTF VLEVMGRHCG YLALVSALAS GADWLFIPEA PPEDGWENFM CERLGETRSR 300
301 GSRLNIIIIA EGAIDRNGKP ISSSYVKDLV VQRLGFDTRV TVLGHVQRGG TPSAFDRILS 360
361 SKMGMEAVMA LLEATPDTPA CVVTLSGNQS VRLPLMECVQ MTKEVQKAMD DKRFDEATQL 420
421 RGGSFENNWN IYKLLAHQKP PKEKSNFSLA ILNVGAPAAG MNAAVRSAVR TGISHGHTVY 480
481 VVHDGFEGLA KGQVQEVGWH DVAGWLGRGG SMLGTKRTLP KGQLESIVEN IRIYGIHALL 540
541 VVGGFEAYEG VLQLVEARGR YEELCIVMCV IPATISNNVP GTDFSLGSDT AVNAAMESCD 600
601 RIKQSASGTK RRVFIVETMG GYCGYLATVT GIAVGADAAY VFEDPFNIHD LKVNVEHMTE 660
661 KMKTDIQRGL VLRNEKCHDY YTTEFLYNLY SSEGKGVFDC RTNVLGHLQQ GGAPTPFDRN 720
721 YGTKLGVKAM LWLSEKLREV YRKGRVFANA PDSACVIGLK KKAVAFSPVT ELKKDTDFEH 780
781 RMPREQWWLS LRLMLKMLAQ YRISMAAYVS GELEHVTRRT LSMDKGF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
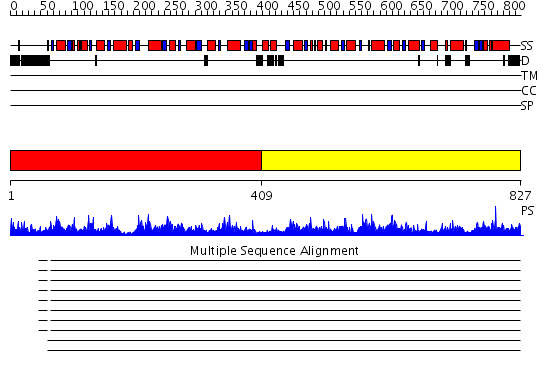
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..408] | 107.0 | ATP-dependent phosphofructokinase | |
| 2 | View Details | [409..827] | 74.154902 | No description for 2higA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)