













| General Information: |
|
| Name(s) found: |
POLA2
[HGNC (HUGO)]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 598 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASAQQLAE ELQIFGLDCE EALIEKLVEL CVQYGQNEEG MVGELIAFCT STHKVGLTSE 60
61 ILNSFEHEFL SKRLSKARHS TCKDSGHAGA RDIVSIQELI EVEEEEEILL NSYTTPSKGS 120
121 QKRAISTPET PLTKRSVSTR SPHQLLSPSS FSPSATPSQK YNSRSNRGEV VTSFGLAQGV 180
181 SWSGRGGAGN ISLKVLGCPE ALTGSYKSMF QKLPDIREVL TCKIEELGSE LKEHYKIEAF 240
241 TPLLAPAQEP VTLLGQIGCD SNGKLNNKSV ILEGDREHSS GAQIPVDLSE LKEYSLFPGQ 300
301 VVIMEGINTT GRKLVATKLY EGVPLPFYQP TEEDADFEQS MVLVACGPYT TSDSITYDPL 360
361 LDLIAVINHD RPDVCILFGP FLDAKHEQVE NCLLTSPFED IFKQCLRTII EGTRSSGSHL 420
421 VFVPSLRDVH HEPVYPQPPF SYSDLSREDK KQVQFVSEPC SLSINGVIFG LTSTDLLFHL 480
481 GAEEISSSSG TSDRFSRILK HILTQRSYYP LYPPQEDMAI DYESFYVYAQ LPVTPDVLII 540
541 PSELRYFVKD VLGCVCVNPG RLTKGQVGGT FARLYLRRPA ADGAERQSPC IAVQVVRI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
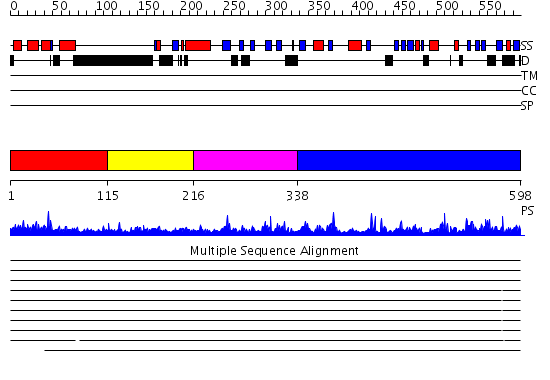
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | 1.055994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [115..215] | 1.135995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [216..337] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [338..598] | 7.0 | No description for 2dxlA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)